[English] 日本語
 Yorodumi
Yorodumi- EMDB-4461: High resolution electron cryo-microscopy structure of the bacteri... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4461 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | High resolution electron cryo-microscopy structure of the bacteriophage PR772 | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Phage / Tectiviridae / Membrane / double-barrel / helix turn helix / helix with a kink / beta-propeller / heteropentamer / penton / VIRUS | ||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||
| Biological species |  Enterobacteria phage PR772 (virus) Enterobacteria phage PR772 (virus) | ||||||||||||
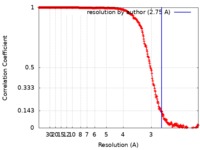
| Method | single particle reconstruction / cryo EM / Resolution: 2.75 Å | ||||||||||||
 Authors Authors | Narayana Reddy HK / Svenda M | ||||||||||||
| Funding support |  Sweden, 3 items Sweden, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Electron cryo-microscopy of bacteriophage PR772 reveals the elusive vertex complex and the capsid architecture. Authors: Hemanth Kn Reddy / Marta Carroni / Janos Hajdu / Martin Svenda /   Abstract: Bacteriophage PR772, a member of the family, has a 70 nm diameter icosahedral protein capsid that encapsulates a lipid membrane, dsDNA, and various internal proteins. An icosahedrally averaged ...Bacteriophage PR772, a member of the family, has a 70 nm diameter icosahedral protein capsid that encapsulates a lipid membrane, dsDNA, and various internal proteins. An icosahedrally averaged CryoEM reconstruction of the wild-type virion and a localized reconstruction of the vertex region reveal the composition and the structure of the vertex complex along with new protein conformations that play a vital role in maintaining the capsid architecture of the virion. The overall resolution of the virion is 2.75 Å, while the resolution of the protein capsid is 2.3 Å. The conventional penta-symmetron formed by the capsomeres is replaced by a large vertex complex in the pseudo T = 25 capsid. All the vertices contain the host-recognition protein, P5; two of these vertices show the presence of the receptor-binding protein, P2. The 3D structure of the vertex complex shows interactions with the viral membrane, indicating a possible mechanism for viral infection. #1:  Journal: Biorxiv / Year: 2019 Journal: Biorxiv / Year: 2019Title: Electron cryo-microscopy of Bacteriophage PR772 reveals the composition and structure of the elusive vertex complex and the capsid architecture Authors: Narayana Reddy HK / Hajdu J / Carroni M / Svenda M | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4461.map.gz emd_4461.map.gz | 2 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4461-v30.xml emd-4461-v30.xml emd-4461.xml emd-4461.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4461_fsc.xml emd_4461_fsc.xml | 30 KB | Display |  FSC data file FSC data file |
| Images |  emd_4461.png emd_4461.png | 335.9 KB | ||
| Filedesc metadata |  emd-4461.cif.gz emd-4461.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4461 http://ftp.pdbj.org/pub/emdb/structures/EMD-4461 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4461 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4461 | HTTPS FTP |
-Related structure data
| Related structure data |  6q5uMC  4462C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4461.map.gz / Format: CCP4 / Size: 2.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4461.map.gz / Format: CCP4 / Size: 2.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bacteriophage PR772
| Entire | Name: Bacteriophage PR772 |
|---|---|
| Components |
|
-Supramolecule #1: Bacteriophage PR772
| Supramolecule | Name: Bacteriophage PR772 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Wild type |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage PR772 (virus) Enterobacteria phage PR772 (virus) |
| Molecular weight | Theoretical: 86 MDa |
-Macromolecule #1: Major Capsid Protein (P3)
| Macromolecule | Name: Major Capsid Protein (P3) / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage PR772 (virus) Enterobacteria phage PR772 (virus) |
| Molecular weight | Theoretical: 43.505492 KDa |
| Sequence | String: MAQVQQLTPA QQAALRNQQA MAANLQARQI VLQQSYPVIQ QVETQTFDPA NRSVFDVTPA NVGIVKGFLV KVTAAIKNNH ATEAVALTD FGPANLVQRV IYYDPDNQRH TETSGWHLHF VNTAKQGAPF LSSMVTDSPI KYGDVMNVID APATIAAGAT G ELTMYYWV ...String: MAQVQQLTPA QQAALRNQQA MAANLQARQI VLQQSYPVIQ QVETQTFDPA NRSVFDVTPA NVGIVKGFLV KVTAAIKNNH ATEAVALTD FGPANLVQRV IYYDPDNQRH TETSGWHLHF VNTAKQGAPF LSSMVTDSPI KYGDVMNVID APATIAAGAT G ELTMYYWV PLAYSETDLT GAVLANVPQS KQRLKLEFAN NNTAFAAVGA NPLEAIYQGA GAADCEFEEI SYTVYQSYLD QL PVGQNGY ILPLIDLSTL YNLENSAQAG LTPNVDFVVQ YANLYRYLST IAVFDNGGSF NAGTDINYLS QRTANFSDTR KLD PKTWAA QTRRRIATDF PKGVYYCDNR DKPIYTLQYG NVGFVVNPKT VNQNARLLMG YEYFTSRTEL VNAGTISTT UniProtKB: Major capsid protein |
-Macromolecule #2: Minor Capsid Protein (P30)
| Macromolecule | Name: Minor Capsid Protein (P30) / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage PR772 (virus) Enterobacteria phage PR772 (virus) |
| Molecular weight | Theoretical: 9.275559 KDa |
| Sequence | String: MALINPQFPY AGPVPIPGPA PTETMPLLNY RVEGRIAGIQ QARQFMPFLQ GPHREVAEQT YYAIGTGIQM GQTFNQPLIN TQEG UniProtKB: Minor capsid protein |
-Macromolecule #3: Infectivity Protein (P16)
| Macromolecule | Name: Infectivity Protein (P16) / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage PR772 (virus) Enterobacteria phage PR772 (virus) |
| Molecular weight | Theoretical: 12.616262 KDa |
| Sequence | String: MDKKKLLYWV GGGLVLIIIW LWFRNRPAAQ VASNWEGPPY MTYNQPQAGS VTLPVAGYTS PSLTLPNRNR SCGCNPAVSA AMAQGADLA SKLTESISSQ LNNYAESLND YLASQAGV UniProtKB: Infectivity protein |
-Macromolecule #4: Spike protein
| Macromolecule | Name: Spike protein / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage PR772 (virus) Enterobacteria phage PR772 (virus) |
| Molecular weight | Theoretical: 34.483898 KDa |
| Sequence | String: MANQQIGGST VTYNGAIPMG GPVAINSVIE IAGTEVLVDL KLDYATGKIS GVQTLYIDLR DFLGDVTVTM PDTGQRITAR AGTQGYYPV LSTNLMKFIV SATIDGKFPM NFINFPIALG VWPSGIKGDK GDPGAPGPAG GTVVVEDSGA SFGESLLDTT S EPGKILVK ...String: MANQQIGGST VTYNGAIPMG GPVAINSVIE IAGTEVLVDL KLDYATGKIS GVQTLYIDLR DFLGDVTVTM PDTGQRITAR AGTQGYYPV LSTNLMKFIV SATIDGKFPM NFINFPIALG VWPSGIKGDK GDPGAPGPAG GTVVVEDSGA SFGESLLDTT S EPGKILVK RISGGSGITV TDYGDEVEIE ASGGGGGGGG VTDALSLMYS TSTGGPASIA ANALTDFDLS GALTVNTVGT GL TKSAAGI QLAAGKSGLY QITMTVKNNT VTTGNYLLRV KYGSSDFVVA CPASSLTAGG TISLLIYCDV LGVPSLDVLK FSL CNDGAA LSNYIINITA AKIN UniProtKB: Spike |
-Macromolecule #5: Penton protein
| Macromolecule | Name: Penton protein / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage PR772 (virus) Enterobacteria phage PR772 (virus) |
| Molecular weight | Theoretical: 13.757388 KDa |
| Sequence | String: MNVNNPNQMT VTPVYNGCDS GEGPQSVRGY FDAVAGENVK YDLTYLADTQ GFTGVQCIYI DNAENDGAFE IDVEETGQRI KCPAGKQGY FPLLVPGRAK FVARHLGSGK KSVPLFFLNF TIAQGVW UniProtKB: Penton |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: C-flat-2/2 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Frames/image: 3-40 / Number grids imaged: 1 / Number real images: 3200 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Software | Name:  Coot (ver. 0.8.9.1) Coot (ver. 0.8.9.1) |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 104.93 / Target criteria: Cross-correlation coefficient |
| Output model |  PDB-6q5u: |
 Movie
Movie Controller
Controller

















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)