[English] 日本語
 Yorodumi
Yorodumi- EMDB-44554: Streptomyces griseus Family 2B encapsulin shell with 2-methylisob... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Streptomyces griseus Family 2B encapsulin shell with 2-methylisoborneol synthase cargo | |||||||||
 Map data Map data | Streptomyces griseus Family 2B encapsulin shell with 2-methylisoborneol synthase cargo | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Encapsulin / nanocompartment / VIRUS LIKE PARTICLE | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Streptomyces griseus subsp. griseus (bacteria) Streptomyces griseus subsp. griseus (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.02 Å | |||||||||
 Authors Authors | Andreas MP / Giessen TW | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: The biosynthesis of the odorant 2-methylisoborneol is compartmentalized inside a protein shell. Authors: Michael P Andreas / Tobias W Giessen /  Abstract: Terpenoids are the largest class of natural products, found across all domains of life. One of the most abundant bacterial terpenoids is the volatile odorant 2-methylisoborneol (2-MIB), partially ...Terpenoids are the largest class of natural products, found across all domains of life. One of the most abundant bacterial terpenoids is the volatile odorant 2-methylisoborneol (2-MIB), partially responsible for the earthy smell of soil and musty taste of contaminated water. Many bacterial 2-MIB biosynthetic gene clusters were thought to encode a conserved transcription factor, named EshA in the model soil bacterium Streptomyces griseus. Here, we revise the function of EshA, now referred to as Sg Enc, and show that it is a Family 2B encapsulin shell protein. Using cryo-electron microscopy, we find that Sg Enc forms an icosahedral protein shell and encapsulates 2-methylisoborneol synthase (2-MIBS) as a cargo protein. Sg Enc contains a cyclic adenosine monophosphate (cAMP) binding domain (CBD)-fold insertion and a unique metal-binding domain, both displayed on the shell exterior. We show that Sg Enc CBDs do not bind cAMP. We find that 2-MIBS cargo loading is mediated by an N-terminal disordered cargo-loading domain and that 2-MIBS activity and Sg Enc shell structure are not modulated by cAMP. Our work redefines the function of EshA and establishes Family 2B encapsulins as cargo-loaded protein nanocompartments involved in secondary metabolite biosynthesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44554.map.gz emd_44554.map.gz | 307.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44554-v30.xml emd-44554-v30.xml emd-44554.xml emd-44554.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_44554_fsc.xml emd_44554_fsc.xml | 14.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_44554.png emd_44554.png | 159.6 KB | ||
| Filedesc metadata |  emd-44554.cif.gz emd-44554.cif.gz | 6.9 KB | ||
| Others |  emd_44554_additional_1.map.gz emd_44554_additional_1.map.gz emd_44554_half_map_1.map.gz emd_44554_half_map_1.map.gz emd_44554_half_map_2.map.gz emd_44554_half_map_2.map.gz | 307.1 MB 301.7 MB 301.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44554 http://ftp.pdbj.org/pub/emdb/structures/EMD-44554 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44554 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44554 | HTTPS FTP |
-Related structure data
| Related structure data |  9bhvMC  9bhuC  9bi0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_44554.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44554.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Streptomyces griseus Family 2B encapsulin shell with 2-methylisoborneol synthase cargo | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.91 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: This map is a homogenous reconstruction of a...
| File | emd_44554_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This map is a homogenous reconstruction of a 3D class from 3D classification using a focus mask that includes 2 cyclic-nucleotide binding domains with C1 symmetry. | ||||||||||||
| Projections & Slices |
| ||||||||||||
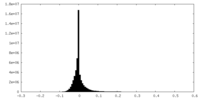
| Density Histograms |
-Half map: Half Map A
| File | emd_44554_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
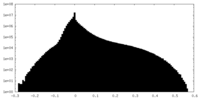
| Density Histograms |
-Half map: Half Map B
| File | emd_44554_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
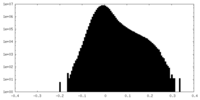
| Density Histograms |
- Sample components
Sample components
-Entire : Streptomyces griseus Family 2B encapsulin shell with 2-methylisob...
| Entire | Name: Streptomyces griseus Family 2B encapsulin shell with 2-methylisoborneol synthase cargo |
|---|---|
| Components |
|
-Supramolecule #1: Streptomyces griseus Family 2B encapsulin shell with 2-methylisob...
| Supramolecule | Name: Streptomyces griseus Family 2B encapsulin shell with 2-methylisoborneol synthase cargo type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Streptomyces griseus subsp. griseus (bacteria) Streptomyces griseus subsp. griseus (bacteria) |
-Supramolecule #2: 2-Methylisoborneol synthase cargo protein
| Supramolecule | Name: 2-Methylisoborneol synthase cargo protein / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Streptomyces griseus subsp. griseus (bacteria) Streptomyces griseus subsp. griseus (bacteria) |
-Macromolecule #1: Nucleotide-binding protein
| Macromolecule | Name: Nucleotide-binding protein / type: protein_or_peptide / ID: 1 Details: Residues 1-39, 147-152, and 216-245 are not resolved in the map Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptomyces griseus subsp. griseus (bacteria) Streptomyces griseus subsp. griseus (bacteria) |
| Molecular weight | Theoretical: 52.014137 KDa |
| Recombinant expression | Organism:  Streptomyces coelicolor A3(2) (bacteria) Streptomyces coelicolor A3(2) (bacteria) |
| Sequence | String: MTVDSTSEAR LEVPRQSSLG TAAARNLAST TKSAPQMQEI TSRWLLRMLP WVETKGGAYR VNRRLTFTVG DGRVEFVQDG STVRVIPQE LGELALLRDF DDAEVLSAIA DRCVQRDFRA GETLVERGTA ADELHLIAHG RIGQASAGSY GDEVTLDVLA D GDRFGEHA ...String: MTVDSTSEAR LEVPRQSSLG TAAARNLAST TKSAPQMQEI TSRWLLRMLP WVETKGGAYR VNRRLTFTVG DGRVEFVQDG STVRVIPQE LGELALLRDF DDAEVLSAIA DRCVQRDFRA GETLVERGTA ADELHLIAHG RIGQASAGSY GDEVTLDVLA D GDRFGEHA LLDEDARWSQ TATAETSGTL LTLSRADFAA VVANSPALRS HLAAFTARSE QRQNHRGEAE IAMSAGHVGE HE LPGAFAD YELKPREYEL SVAQTILRVH TRVADLYNGP MNQTEEQLRL TIEALRERQE HELINNREFG LLHNADFKQR IQT HSGPPT PDDLDELLCR RRGTKFFLAH PRTIAAMGRE FNARGLYPDH TDLGGQQVPA WRGVPILPCG KIPITPERTS SILA LRTGE EDQGVIGLRQ TGLPDEYEPG LSVRFMNIDE KAIISYLVST YYSAAILVPD AVGVLENVQI ANWPR UniProtKB: Sporulation protein |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.0 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 150 mM NaCl, 20 mM Tris pH 7.5 | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Details: 60 seconds at 5 mA under vacuum | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV / Details: Blot force: 20 Blot time: 4 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Number grids imaged: 1 / Average exposure time: 8.0 sec. / Average electron dose: 42.94 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 45000 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Details | An AlphaFold-generated model was fit into the map using ChimeraX v1.5. The model was then refined iteratively by alternating rounds of manual refinement in Coot v8.9.1 and real-space refinements using PHENIX v1.20.1-4487. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 115.8 / Target criteria: cross-correlation coefficient |
| Output model |  PDB-9bhv: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)