[English] 日本語
 Yorodumi
Yorodumi- EMDB-44311: Cryo-EM structure of native SWR1, free complex (consensus map fil... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of native SWR1, free complex (consensus map filtered by local resolution) | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Chromatin Remodeler / Snf2 family ATPase / histone exchange / H2A.Z / GENE REGULATION | ||||||||||||
| Biological species |  | ||||||||||||
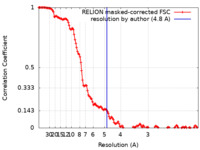
| Method | single particle reconstruction / cryo EM / Resolution: 4.8 Å | ||||||||||||
 Authors Authors | Louder RK / Park G / Wu C | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Molecular basis of global promoter sensing and nucleosome capture by the SWR1 chromatin remodeler. Authors: Robert K Louder / Giho Park / Ziyang Ye / Justin S Cha / Anne M Gardner / Qin Lei / Anand Ranjan / Eva Höllmüller / Florian Stengel / B Franklin Pugh / Carl Wu /   Abstract: The SWR1 chromatin remodeling complex is recruited to +1 nucleosomes downstream of transcription start sites of eukaryotic promoters, where it exchanges histone H2A for the specialized variant H2A.Z. ...The SWR1 chromatin remodeling complex is recruited to +1 nucleosomes downstream of transcription start sites of eukaryotic promoters, where it exchanges histone H2A for the specialized variant H2A.Z. Here, we use cryoelectron microscopy (cryo-EM) to resolve the structural basis of the SWR1 interaction with free DNA, revealing a distinct open conformation of the Swr1 ATPase that enables sliding from accessible DNA to nucleosomes. A complete structural model of the SWR1-nucleosome complex illustrates critical roles for Swc2 and Swc3 subunits in oriented nucleosome engagement by SWR1. Moreover, an extended DNA-binding α helix within the Swc3 subunit enables sensing of nucleosome linker length and is essential for SWR1-promoter-specific recruitment and activity. The previously unresolved N-SWR1 subcomplex forms a flexible extended structure, enabling multivalent recognition of acetylated histone tails by reader domains to further direct SWR1 toward the +1 nucleosome. Altogether, our findings provide a generalizable mechanism for promoter-specific targeting of chromatin and transcription complexes. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44311.map.gz emd_44311.map.gz | 124.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44311-v30.xml emd-44311-v30.xml emd-44311.xml emd-44311.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_44311_fsc.xml emd_44311_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_44311.png emd_44311.png | 117 KB | ||
| Masks |  emd_44311_msk_1.map emd_44311_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-44311.cif.gz emd-44311.cif.gz | 4.5 KB | ||
| Others |  emd_44311_half_map_1.map.gz emd_44311_half_map_1.map.gz emd_44311_half_map_2.map.gz emd_44311_half_map_2.map.gz | 171.5 MB 171.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44311 http://ftp.pdbj.org/pub/emdb/structures/EMD-44311 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44311 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44311 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_44311.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44311.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||
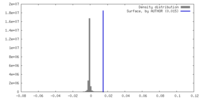
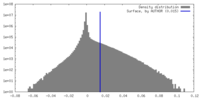
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_44311_msk_1.map emd_44311_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_44311_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_44311_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Native SWR1 complex
| Entire | Name: Native SWR1 complex |
|---|---|
| Components |
|
-Supramolecule #1: Native SWR1 complex
| Supramolecule | Name: Native SWR1 complex / type: complex / ID: 1 / Parent: 0 / Details: Endogenously purified yeast SWR1 complex |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.19 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7.6 Details: 25 mM HEPES, 0.2 mM EDTA, 1 mM MgCl2, 100 mM KOAc, 1 mM DTT, 0.25 mM TCEP, 1% glycerol buffer |
| Grid | Model: Quantifoil R0.6/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 90 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 3 second blot time and blot force of 5.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 3691 / Average exposure time: 4.0 sec. / Average electron dose: 54.0 e/Å2 Details: Each micrograph was fractionated into 64 frames within a 4 second exposure. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 48543 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.7 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)