[English] 日本語
 Yorodumi
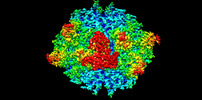
Yorodumi- EMDB-44273: Fab1-3 in complex with the capsid of Adeno-associated virus type 9 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Fab1-3 in complex with the capsid of Adeno-associated virus type 9 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Adeno-associated virus / AAV / capsid / Fab-complex / antibody / NAb / VIRUS / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology | Phospholipase A2-like domain / Phospholipase A2-like domain / Parvovirus coat protein VP2 / Parvovirus coat protein VP1/VP2 / Parvovirus coat protein VP1/VP2 / Capsid/spike protein, ssDNA virus / T=1 icosahedral viral capsid / structural molecule activity / Capsid protein VP1 Function and homology information Function and homology information | |||||||||
| Biological species |   Adeno-associated virus / Adeno-associated virus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.13 Å | |||||||||
 Authors Authors | Mietzsch M / McKenna R | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Structural characterization of antibody-responses following Zolgensma treatment for AAV capsid engineering to expand patient cohorts. Authors: Mario Mietzsch / Jane Hsi / Austin R Nelson / Neeta Khandekar / Ann-Maree Huang / Nicholas Jc Smith / Jon Zachary / Lindsay Potts / Michelle A Farrar / Paul Chipman / Mohammad Ghanem / Ian E ...Authors: Mario Mietzsch / Jane Hsi / Austin R Nelson / Neeta Khandekar / Ann-Maree Huang / Nicholas Jc Smith / Jon Zachary / Lindsay Potts / Michelle A Farrar / Paul Chipman / Mohammad Ghanem / Ian E Alexander / Grant J Logan / Juha T Huiskonen / Robert McKenna /    Abstract: Monoclonal antibodies are useful tools to dissect the neutralizing antibody response against the adeno-associated virus (AAV) capsids that are used as gene therapy delivery vectors. The presence of ...Monoclonal antibodies are useful tools to dissect the neutralizing antibody response against the adeno-associated virus (AAV) capsids that are used as gene therapy delivery vectors. The presence of pre-existing neutralizing antibodies in large portions of the human population poses a significant challenge for AAV-mediated gene therapy, primarily targeting the capsid leading to vector inactivation and loss of treatment efficacy. This study structurally characterizes the interactions of 21 human-derived neutralizing antibodies from three patients treated with the AAV9 vector, Zolgensma®, utilizing high-resolution cryo-electron microscopy. The antibodies bound to the 2-fold depression or the 3-fold protrusions do not conform to the icosahedral symmetry of the capsid, thus requiring localized reconstructions. These complex structures provide unprecedented details of the mAbs binding interfaces, with many antibodies inducing structural perturbations of the capsid upon binding. Key surface capsid amino acid residues were identified facilitating the design of capsid variants with antibody escape phenotypes. These AAV9 capsid variants have the potential to expand the patient cohort to include those that were previously excluded due to their pre-existing neutralizing antibodies against the wtAAV9 capsid, and the possibly of further treatment to those requiring redosing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44273.map.gz emd_44273.map.gz | 28.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44273-v30.xml emd-44273-v30.xml emd-44273.xml emd-44273.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_44273.png emd_44273.png | 71.5 KB | ||
| Filedesc metadata |  emd-44273.cif.gz emd-44273.cif.gz | 6.6 KB | ||
| Others |  emd_44273_half_map_1.map.gz emd_44273_half_map_1.map.gz emd_44273_half_map_2.map.gz emd_44273_half_map_2.map.gz | 23 MB 23 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44273 http://ftp.pdbj.org/pub/emdb/structures/EMD-44273 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44273 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44273 | HTTPS FTP |
-Related structure data
| Related structure data |  9b6pMC  9b6nC  9b6oC  9b6qC  9b6rC  9b6sC  9b6tC  9b7kC  9b7lC  9b7mC  9b7nC  9b7oC  9b7pC  9b7qC  9b7rC  9b7sC  9b7tC  9b7uC  9b7vC  9b7wC  9b7xC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_44273.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44273.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_44273_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_44273_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Adeno-associated virus
| Entire | Name:   Adeno-associated virus Adeno-associated virus |
|---|---|
| Components |
|
-Supramolecule #1: Adeno-associated virus
| Supramolecule | Name: Adeno-associated virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 272636 / Sci species name: Adeno-associated virus / Virus type: VIRION / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: No |
|---|---|
| Virus shell | Shell ID: 1 / T number (triangulation number): 1 |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Adeno-associated virus / Strain: AAV9 Adeno-associated virus / Strain: AAV9 |
| Molecular weight | Theoretical: 59.873875 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASGGGAPVA DNNEGADGVG SSSGNWHCDS QWLGDRVITT STRTWALPTY NNHLYKQISN STSGGSSNDN AYFGYSTPWG YFDFNRFHC HFSPRDWQRL INNNWGFRPK RLNFKLFNIQ VKEVTDNNGV KTIANNLTST VQVFTDSDYQ LPYVLGSAHE G CLPPFPAD ...String: MASGGGAPVA DNNEGADGVG SSSGNWHCDS QWLGDRVITT STRTWALPTY NNHLYKQISN STSGGSSNDN AYFGYSTPWG YFDFNRFHC HFSPRDWQRL INNNWGFRPK RLNFKLFNIQ VKEVTDNNGV KTIANNLTST VQVFTDSDYQ LPYVLGSAHE G CLPPFPAD VFMIPQYGYL TLNDGSQAVG RSSFYCLEYF PSQMLRTGNN FQFSYEFENV PFHSSYAHSQ SLDRLMNPLI DQ YLYYLSK TINGSGQNQQ TLKFSVAGPS NMAVQGRNYI PGPSYRQQRV STTVTQNNNS EFAWPGASSW ALNGRNSLMN PGP AMASHK EGEDRFFPLS GSLIFGKQGT GRDNVDADKV MITNEEEIKT TNPVATESYG QVATNHQSAQ AQAQTGWVQN QGIL PGMVW QDRDVYLQGP IWAKIPHTDG NFHPSPLMGG FGMKHPPPQI LIKNTPVPAD PPTAFNKDKL NSFITQYSTG QVSVE IEWE LQKENSKRWN PEIQYTSNYY KSNNVEFAVN TEGVYSEPRP IGTRYLTRNL UniProtKB: Capsid protein VP1 |
-Macromolecule #2: Fab1-3 heavy chain
| Macromolecule | Name: Fab1-3 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.089645 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VQLVESGGGV VQPGRSLRLS CAASGFSFSS FVFHWVRQAP GKGLEWVAVI SYDGSNKNYA DSVKGRFTIS RDNSKNTLFL QMNSLSADD TAVYYCARGA ICSSTTCYLG DSYYFYGMDV WGQGTTVTVS S |
-Macromolecule #3: Fab1-3 light chain
| Macromolecule | Name: Fab1-3 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.244273 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LTQPPSVSGA PGQRVTISCT GSRSNIGAGY DVHWYQQLPG TAPKLLIYGN SNRPSGVPDR FSGSKSGTSA SLAITGLQTD DEADYYCQS YDSSLSGSVF GGGTKLTVLG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)