[English] 日本語
 Yorodumi
Yorodumi- EMDB-43519: Structure of the gamma tubulin ring complex nucleated microtubule... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the gamma tubulin ring complex nucleated microtubule protofilament. | |||||||||
 Map data Map data | Structure of the gamma tubulin ring complex nucleated microtubule. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Microtubule / CELL CYCLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationnetrin receptor binding / Post-chaperonin tubulin folding pathway / dorsal root ganglion development / Cilium Assembly / cytoskeleton-dependent intracellular transport / Carboxyterminal post-translational modifications of tubulin / Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Sealing of the nuclear envelope (NE) by ESCRT-III / Intraflagellar transport / Formation of tubulin folding intermediates by CCT/TriC ...netrin receptor binding / Post-chaperonin tubulin folding pathway / dorsal root ganglion development / Cilium Assembly / cytoskeleton-dependent intracellular transport / Carboxyterminal post-translational modifications of tubulin / Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Sealing of the nuclear envelope (NE) by ESCRT-III / Intraflagellar transport / Formation of tubulin folding intermediates by CCT/TriC / Gap junction assembly / Prefoldin mediated transfer of substrate to CCT/TriC / Kinesins / Assembly and cell surface presentation of NMDA receptors / COPI-independent Golgi-to-ER retrograde traffic / COPI-dependent Golgi-to-ER retrograde traffic / Recycling pathway of L1 / RHOH GTPase cycle / microtubule-based process / RHO GTPases activate IQGAPs / Hedgehog 'off' state / intercellular bridge / COPI-mediated anterograde transport / cytoplasmic microtubule / Activation of AMPK downstream of NMDARs / peptide binding / MHC class II antigen presentation / Recruitment of NuMA to mitotic centrosomes / Mitotic Prometaphase / cellular response to interleukin-4 / EML4 and NUDC in mitotic spindle formation / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / axon guidance / Resolution of Sister Chromatid Cohesion / cell periphery / Translocation of SLC2A4 (GLUT4) to the plasma membrane / filopodium / RHO GTPases Activate Formins / PKR-mediated signaling / structural constituent of cytoskeleton / microtubule cytoskeleton organization / HCMV Early Events / Aggrephagy / The role of GTSE1 in G2/M progression after G2 checkpoint / mitotic spindle / Separation of Sister Chromatids / mitotic cell cycle / lamellipodium / double-stranded RNA binding / microtubule cytoskeleton / growth cone / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / cilium / axon / cell division / neuronal cell body / GTPase activity / ubiquitin protein ligase binding / dendrite / GTP binding / structural molecule activity / extracellular exosome / metal ion binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.66 Å | |||||||||
 Authors Authors | Aher A / Urnavicius L / Kapoor TM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structure of the γ-tubulin ring complex-capped microtubule. Authors: Amol Aher / Linas Urnavicius / Allen Xue / Kasahun Neselu / Tarun M Kapoor /  Abstract: Microtubules are composed of α-tubulin and β-tubulin dimers positioned head-to-tail to form protofilaments that associate laterally in varying numbers. It is not known how cellular microtubules ...Microtubules are composed of α-tubulin and β-tubulin dimers positioned head-to-tail to form protofilaments that associate laterally in varying numbers. It is not known how cellular microtubules assemble with the canonical 13-protofilament architecture, resulting in micrometer-scale α/β-tubulin tracks for intracellular transport that align with, rather than spiral along, the long axis of the filament. We report that the human ~2.3 MDa γ-tubulin ring complex (γ-TuRC), an essential regulator of microtubule formation that contains 14 γ-tubulins, selectively nucleates 13-protofilament microtubules. Cryogenic electron microscopy reconstructions of γ-TuRC-capped microtubule minus ends reveal the extensive intra-domain and inter-domain motions of γ-TuRC subunits that accommodate luminal bridge components and establish lateral and longitudinal interactions between γ-tubulins and α-tubulins. Our structures suggest that γ-TuRC, an inefficient nucleation template owing to its splayed conformation, can transform into a compacted cap at the microtubule minus end and set the lattice architecture of cellular microtubules. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43519.map.gz emd_43519.map.gz | 41.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43519-v30.xml emd-43519-v30.xml emd-43519.xml emd-43519.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43519_fsc.xml emd_43519_fsc.xml | 16.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_43519.png emd_43519.png | 24.9 KB | ||
| Masks |  emd_43519_msk_1.map emd_43519_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-43519.cif.gz emd-43519.cif.gz | 6.8 KB | ||
| Others |  emd_43519_half_map_1.map.gz emd_43519_half_map_1.map.gz emd_43519_half_map_2.map.gz emd_43519_half_map_2.map.gz | 474.5 MB 474.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43519 http://ftp.pdbj.org/pub/emdb/structures/EMD-43519 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43519 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43519 | HTTPS FTP |
-Related structure data
| Related structure data |  8vt7MC  8va2C  8vrdC  8vrjC  8vrkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43519.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43519.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the gamma tubulin ring complex nucleated microtubule. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1769 Å | ||||||||||||||||||||||||||||||||||||
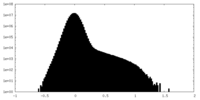
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_43519_msk_1.map emd_43519_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
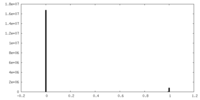
| Density Histograms |
-Half map: Structure of the gamma tubulin ring complex nucleated microtubule.
| File | emd_43519_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the gamma tubulin ring complex nucleated microtubule. | ||||||||||||
| Projections & Slices |
| ||||||||||||
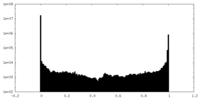
| Density Histograms |
-Half map: Structure of the gamma tubulin ring complex nucleated microtubule.
| File | emd_43519_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the gamma tubulin ring complex nucleated microtubule. | ||||||||||||
| Projections & Slices |
| ||||||||||||
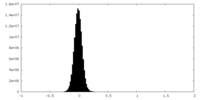
| Density Histograms |
- Sample components
Sample components
-Entire : Gamma tubulin ring complex nucleated microtubule
| Entire | Name: Gamma tubulin ring complex nucleated microtubule |
|---|---|
| Components |
|
-Supramolecule #1: Gamma tubulin ring complex nucleated microtubule
| Supramolecule | Name: Gamma tubulin ring complex nucleated microtubule / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2, #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Tubulin beta-3 chain
| Macromolecule | Name: Tubulin beta-3 chain / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.276367 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPSGNYVGDS DLQLERISVY YNEASSHKYV PRAILVDLEP GTMDSVRSGA FGHLFRPDN FIFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKECENCDC LQGFQLTHSL GGGTGSGMGT LLISKVREEY P DRIMNTFS ...String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPSGNYVGDS DLQLERISVY YNEASSHKYV PRAILVDLEP GTMDSVRSGA FGHLFRPDN FIFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKECENCDC LQGFQLTHSL GGGTGSGMGT LLISKVREEY P DRIMNTFS VVPSPKVSDT VVEPYNATLS IHQLVENTDE TYCIDNEALY DICFRTLKLA TPTYGDLNHL VSATMSGVTT SL RFPGQLN ADLRKLAVNM VPFPRLHFFM PGFAPLTARG SQQYRALTVP ELTQQMFDAK NMMAACDPRH GRYLTVATVF RGR MSMKEV DEQMLAIQSK NSSYFVEWIP NNVKVAVCDI PPRGLKMSST FIGNSTAIQE LFKRISEQFT AMFRRKAFLH WYTG EGMDE MEFTEAESNM NDLVSEYQQY QDATAEEEGE MYEDDEEESE AQGPKENLYF Q UniProtKB: Tubulin beta-3 chain |
-Macromolecule #2: Tubulin alpha-1B chain
| Macromolecule | Name: Tubulin alpha-1B chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.019297 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIHHHHHHGG GDDSFNTFFS ETGAGKHVPR AVFVDLEPTV IDEVRTGTY RQLFHPEQLI TGKEDAANNY ARGHYTIGKE IIDLVLDRIR KLADQCTGLQ GFLVFHSFGG GTGSGFTSLL M ERLSVDYG ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIHHHHHHGG GDDSFNTFFS ETGAGKHVPR AVFVDLEPTV IDEVRTGTY RQLFHPEQLI TGKEDAANNY ARGHYTIGKE IIDLVLDRIR KLADQCTGLQ GFLVFHSFGG GTGSGFTSLL M ERLSVDYG KKSKLEFSIY PAPQVSTAVV EPYNSILTTH TTLEHSDCAF MVDNEAIYDI CRRNLDIERP TYTNLNRLIS QI VSSITAS LRFDGALNVD LTDFQTNLVP YPRIHFPLAT YAPVISAEKA YHEQLSVAEI TNACFEPANQ MVKCDPRHGK YMA CCLLYR GDVVPKDVNA AIATIKTKRS IQFVDWCPTG FKVGINYQPP TVVPGGDLAK VQRAVCMLSN TTAIAEAWAR LDHK FDLMY AKRAFVHWYV GEGMEEGEFS EAREDMAALE KDYEEVGVDS VEGEGEEEGE EY UniProtKB: Tubulin alpha-1B chain |
-Macromolecule #3: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 2 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Macromolecule #4: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 2 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)