+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Density map for free recombinant gamma tubulin ring complex | |||||||||
 Map data Map data | density map for free recombinant gamma tubulin ring complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmicrotubule nucleation by interphase microtubule organizing center / gamma-tubulin complex localization / microtubule nucleator activity / positive regulation of norepinephrine uptake / Regulation of CDH1 Function / Formation of the polybromo-BAF (pBAF) complex / Formation of the non-canonical BAF (ncBAF) complex / polar microtubule / Formation of the canonical BAF (cBAF) complex / Formation of the embryonic stem cell BAF (esBAF) complex ...microtubule nucleation by interphase microtubule organizing center / gamma-tubulin complex localization / microtubule nucleator activity / positive regulation of norepinephrine uptake / Regulation of CDH1 Function / Formation of the polybromo-BAF (pBAF) complex / Formation of the non-canonical BAF (ncBAF) complex / polar microtubule / Formation of the canonical BAF (cBAF) complex / Formation of the embryonic stem cell BAF (esBAF) complex / Formation of neuronal progenitor and neuronal BAF (npBAF and nBAF) / interphase microtubule organizing center / gamma-tubulin complex / gamma-tubulin ring complex / mitotic spindle microtubule / bBAF complex / cellular response to cytochalasin B / meiotic spindle organization / npBAF complex / nBAF complex / brahma complex / regulation of transepithelial transport / morphogenesis of a polarized epithelium / Formation of annular gap junctions / Formation of the dystrophin-glycoprotein complex (DGC) / structural constituent of postsynaptic actin cytoskeleton / GBAF complex / Gap junction degradation / Folding of actin by CCT/TriC / microtubule nucleation / regulation of G0 to G1 transition / Cell-extracellular matrix interactions / protein localization to adherens junction / dense body / Tat protein binding / postsynaptic actin cytoskeleton / Prefoldin mediated transfer of substrate to CCT/TriC / RSC-type complex / gamma-tubulin binding / regulation of double-strand break repair / regulation of nucleotide-excision repair / non-motile cilium / Adherens junctions interactions / RHOF GTPase cycle / adherens junction assembly / apical protein localization / Sensory processing of sound by inner hair cells of the cochlea / Sensory processing of sound by outer hair cells of the cochlea / Interaction between L1 and Ankyrins / tight junction / regulation of mitotic metaphase/anaphase transition / SWI/SNF complex / positive regulation of T cell differentiation / apical junction complex / positive regulation of double-strand break repair / maintenance of blood-brain barrier / regulation of norepinephrine uptake / transporter regulator activity / nitric-oxide synthase binding / pericentriolar material / cortical cytoskeleton / positive regulation of stem cell population maintenance / NuA4 histone acetyltransferase complex / establishment or maintenance of cell polarity / cell leading edge / Recycling pathway of L1 / Regulation of MITF-M-dependent genes involved in pigmentation / microtubule organizing center / mitotic sister chromatid segregation / brush border / regulation of G1/S transition of mitotic cell cycle / EPH-ephrin mediated repulsion of cells / negative regulation of cell differentiation / mitotic spindle assembly / kinesin binding / RHO GTPases Activate WASPs and WAVEs / regulation of synaptic vesicle endocytosis / positive regulation of myoblast differentiation / single fertilization / RHO GTPases activate IQGAPs / regulation of protein localization to plasma membrane / positive regulation of double-strand break repair via homologous recombination / cytoplasmic microtubule / spindle assembly / cytoplasmic microtubule organization / EPHB-mediated forward signaling / cytoskeleton organization / Loss of Nlp from mitotic centrosomes / Loss of proteins required for interphase microtubule organization from the centrosome / Recruitment of mitotic centrosome proteins and complexes / centriole / Recruitment of NuMA to mitotic centrosomes / substantia nigra development / Anchoring of the basal body to the plasma membrane / axonogenesis / AURKA Activation by TPX2 / calyx of Held / nitric-oxide synthase regulator activity / condensed nuclear chromosome / mitotic spindle organization Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
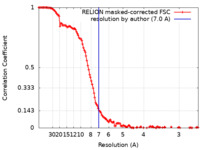
| Method | single particle reconstruction / cryo EM / Resolution: 7.0 Å | |||||||||
 Authors Authors | Aher A / Urnavicius L / Kapoor TM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structure of the γ-tubulin ring complex-capped microtubule. Authors: Amol Aher / Linas Urnavicius / Allen Xue / Kasahun Neselu / Tarun M Kapoor /  Abstract: Microtubules are composed of α-tubulin and β-tubulin dimers positioned head-to-tail to form protofilaments that associate laterally in varying numbers. It is not known how cellular microtubules ...Microtubules are composed of α-tubulin and β-tubulin dimers positioned head-to-tail to form protofilaments that associate laterally in varying numbers. It is not known how cellular microtubules assemble with the canonical 13-protofilament architecture, resulting in micrometer-scale α/β-tubulin tracks for intracellular transport that align with, rather than spiral along, the long axis of the filament. We report that the human ~2.3 MDa γ-tubulin ring complex (γ-TuRC), an essential regulator of microtubule formation that contains 14 γ-tubulins, selectively nucleates 13-protofilament microtubules. Cryogenic electron microscopy reconstructions of γ-TuRC-capped microtubule minus ends reveal the extensive intra-domain and inter-domain motions of γ-TuRC subunits that accommodate luminal bridge components and establish lateral and longitudinal interactions between γ-tubulins and α-tubulins. Our structures suggest that γ-TuRC, an inefficient nucleation template owing to its splayed conformation, can transform into a compacted cap at the microtubule minus end and set the lattice architecture of cellular microtubules. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43481.map.gz emd_43481.map.gz | 24.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43481-v30.xml emd-43481-v30.xml emd-43481.xml emd-43481.xml | 26.9 KB 26.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43481_fsc.xml emd_43481_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_43481.png emd_43481.png | 167.3 KB | ||
| Masks |  emd_43481_msk_1.map emd_43481_msk_1.map | 190.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-43481.cif.gz emd-43481.cif.gz | 9.7 KB | ||
| Others |  emd_43481_half_map_1.map.gz emd_43481_half_map_1.map.gz emd_43481_half_map_2.map.gz emd_43481_half_map_2.map.gz | 150.5 MB 150.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43481 http://ftp.pdbj.org/pub/emdb/structures/EMD-43481 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43481 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43481 | HTTPS FTP |
-Related structure data
| Related structure data |  8vrdMC  8va2C  8vrjC  8vrkC  8vt7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43481.map.gz / Format: CCP4 / Size: 190.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43481.map.gz / Format: CCP4 / Size: 190.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | density map for free recombinant gamma tubulin ring complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_43481_msk_1.map emd_43481_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map for the free recombinant gamma tubulin ring complex
| File | emd_43481_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map for the free recombinant gamma tubulin ring complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map for the density map for free...
| File | emd_43481_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map for the density map for free recombinant gamma tubulin ring complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Gamma tubulin ring complex
| Entire | Name: Gamma tubulin ring complex |
|---|---|
| Components |
|
-Supramolecule #1: Gamma tubulin ring complex
| Supramolecule | Name: Gamma tubulin ring complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #5, #1, #7-#8, #2-#4, #6 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Gamma-tubulin complex component 3
| Macromolecule | Name: Gamma-tubulin complex component 3 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 103.710102 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MATPDQKSPN VLLQNLCCRI LGRSEADVAQ QFQYAVRVIG SNFAPTVERD EFLVAEKIKK ELIRQRREAD AALFSELHRK LHSQGVLKN KWSILYLLLS LSEDPRRQPS KVSSYATLFA QALPRDAHST PYYYARPQTL PLSYQDRSAQ SAQSSGSVGS S GISSIGLC ...String: MATPDQKSPN VLLQNLCCRI LGRSEADVAQ QFQYAVRVIG SNFAPTVERD EFLVAEKIKK ELIRQRREAD AALFSELHRK LHSQGVLKN KWSILYLLLS LSEDPRRQPS KVSSYATLFA QALPRDAHST PYYYARPQTL PLSYQDRSAQ SAQSSGSVGS S GISSIGLC ALSGPAPAPQ SLLPGQSNQA PGVGDCLRQQ LGSRLAWTLT ANQPSSQATT SKGVPSAVSR NMTRSRREGD TG GTMEITE AALVRDILYV FQGIDGKNIK MNNTENCYKV EGKANLSRSL RDTAVRLSEL GWLHNKIRRY TDQRSLDRSF GLV GQSFCA ALHQELREYY RLLSVLHSQL QLEDDQGVNL GLESSLTLRR LLVWTYDPKI RLKTLAALVD HCQGRKGGEL ASAV HAYTK TGDPYMRSLV QHILSLVSHP VLSFLYRWIY DGELEDTYHE FFVASDPTVK TDRLWHDKYT LRKSMIPSFM TMDQS RKVL LIGKSINFLH QVCHDQTPTT KMIAVTKSAE SPQDAADLFT DLENAFQGKI DAAYFETSKY LLDVLNKKYS LLDHMQ AMR RYLLLGQGDF IRHLMDLLKP ELVRPATTLY QHNLTGILET AVRATNAQFD SPEILRRLDV RLLEVSPGDT GWDVFSL DY HVDGPIATVF TRECMSHYLR VFNFLWRAKR MEYILTDIRK GHMCNAKLLR NMPEFSGVLH QCHILASEMV HFIHQMQY Y ITFEVLECSW DELWNKVQQA QDLDHIIAAH EVFLDTIISR CLLDSDSRAL LNQLRAVFDQ IIELQNAQDA IYRAALEEL QRRLQFEEKK KQREIEGQWG VTAAEEEEEN KRIGEFKESI PKMCSQLRIL THFYQGIVQQ FLVLLTTSSD ESLRFLSFRL DFNEHYKAR EPRLRVSLGT RGRRSSHT UniProtKB: Gamma-tubulin complex component 3 |
-Macromolecule #2: TUBGCP6 protein
| Macromolecule | Name: TUBGCP6 protein / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 199.732516 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASITQLFDD LCEALLPAAK THLGQRSVNR KRAKRSLKKV AYNALFTNLF QDETQQLQPD MSKLPARNKI LMLSFDLRVG GLGPKADRL EELVEELEAA PCCPLLEVGS VLDLLVQLAG SGPPQVLPRK RDYFLNNKHV GRNVPYSGYD CDDLSVFEMD V QSLISREE ...String: MASITQLFDD LCEALLPAAK THLGQRSVNR KRAKRSLKKV AYNALFTNLF QDETQQLQPD MSKLPARNKI LMLSFDLRVG GLGPKADRL EELVEELEAA PCCPLLEVGS VLDLLVQLAG SGPPQVLPRK RDYFLNNKHV GRNVPYSGYD CDDLSVFEMD V QSLISREE CLCHSMIQET LQVMEAAPGT GLPTVGLFSF GDPCGDRFER DTRVSLFGAL VHSRTYDMDV RLGLPPVPDN AD LSGLAIK VPPSVDQWED EGFQSASNLT PDSQSEPSVT PDVDLWEAAL TYEASKRRCW ERVGCPPGHR EEPYLTEAGR DAF DKFCRL HQGELQLLAG GVLQAPQPVL VKECELVKDV LNVLIGVVSA TFSLCQPAQA FVVKRGVHVS GASPESISSL LSEV AEYGT CYTRLSHFSL QPVLDSLYSK GLVFQAFTSG LRRYLQYYRA CVLSTPPTLS LLTIGFLFKK LGRQLRYLAE LCGVG AVLP GTCGGGPRAA FPTGVKLLSY LYQEALHNCS NEHYPVLLSL LKTSCEPYTR FIHDWVYSGV FRDAYGEFMI QVNHEY LSF RDKLYWTHGY VLISKEVEDC VPVFLKHIAH DIYVCGKTIN LLKLCCPRHY LCWSDVPVPR ISVIFSLEEL KEIEKDC AV YVGRMERVAR HSSVSKEEKE LRMEIAKQEL IAHAREAASR VLSALSDRQM SERMALDARK REQFQRLKEQ FVKDQERR Q AARQEELDDD FSYARELRDR ERRLKSLEEE LERKASKLSA EAARREQKAL WRIQRHRLES ARLRFLLEDE KHIQEMLKA VSEAHQPQEP PDVLLSVHPQ VTSPGPEHPE GGQGCDSGSA EQHSPAWDGW NRPGLLTPQP LKPLAVGAGG RGLQQAEGAR PFSDSLSIG DFLPVGPGAE PSVQTGMVPL LEVALQTINL DLPPSAPGEA PAAASTQPSR PQEYDFSTVL RPAVATSPAP G PLQAAECS LGSSGLQLWE DSCGKMDACG SASRETLLPS HPPRRAALEE GSSQPTERLF GQVSGGGLPT GDYASEIAPT RP RWNTHGH VSDASIRVGE NVSDVAPTQP RWNTHGHVSN ASISLGESVS DVAPTRPRWN IHGHVSNASI RVGENVSDVA PTR PRWNTH GHVSNASIRV GENVSDVAPT RPRWNTHGHV SDASISLGES VSDMAPARPR WNTHGHVSDA SISLGESVSD MAPT RPRWN THGHVSDTSI RVGENVSDVA PIRSRCNTHG HVSDASISLG EPVSDVVSTR PRWNTHVPIP PPHMVLGALS PEAEP NTPR PQQSPPGHTS QSALSLGAQS AVLDCGPRLP VEVGPSLSSP SSGCGEGSIS VGENVSDVAP TQPWWPNTPG DSVSEE LGP GRSGDTEDLS PNWPLNSQED TAAQSSPGRG EEAEASAAEA QGGEQAYLAG LAGQYHLERY PDSYESMSEP PIAHLLR PV LPRAFAFPVD PQVQSAADET AVQLSELLTL PVLMKRSITA PLAAHISLVN KAAVDYFFVE LHLEAHYEAL RHFLLMED G EFAQSLSDLL FEKLGAGQTP GELLNPLVLN SVLSKALQCS LHGDTPHASN LSLALKYLPE VFAPNAPDVL SCLELRYKV DWPLNIVITE GCLSKYSGVF SFLLQLKLMM WALKDVCFHL KRTALLSHMA GSVQFRQLQL FKHEMQHFVK VIQGYIANQI LHVTWCEFR ARLATVGDLE EIQRAHAEYL HEAVFRGLLT EKAAPVMNVI HSIFSLVLKF RSQLISQAWG PPGGPRGAEH P NFALMQQS YNTFKYYSHF LFKVVTKLVN RGYQPHLEDF LLRINFNNYY QDA UniProtKB: Gamma-tubulin complex component 6 |
-Macromolecule #3: Mitotic-spindle organizing protein 1
| Macromolecule | Name: Mitotic-spindle organizing protein 1 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.485724 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASSSGAGAA AAAAAANLNA VRETMDVLLE ISRILNTGLD METLSICVRL CEQGINPEAL SSVIKELRKA TEALKAAENM TS UniProtKB: Mitotic-spindle organizing protein 1 |
-Macromolecule #4: Actin, cytoplasmic 1
| Macromolecule | Name: Actin, cytoplasmic 1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.78266 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDDDIAALVV DNGSGMCKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS KRGILTLKYP IEHGIVTNWD DMEKIWHHT FYNELRVAPE EHPVLLTEAP LNPKANREKM TQIMFETFNT PAMYVAIQAV LSLYASGRTT GIVMDSGDGV T HTVPIYEG ...String: MDDDIAALVV DNGSGMCKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS KRGILTLKYP IEHGIVTNWD DMEKIWHHT FYNELRVAPE EHPVLLTEAP LNPKANREKM TQIMFETFNT PAMYVAIQAV LSLYASGRTT GIVMDSGDGV T HTVPIYEG YALPHAILRL DLAGRDLTDY LMKILTERGY SFTTTAEREI VRDIKEKLCY VALDFEQEMA TAASSSSLEK SY ELPDGQV ITIGNERFRC PEALFQPSFL GMESCGIHET TFNSIMKCDV DIRKDLYANT VLSGGTTMYP GIADRMQKEI TAL APSTMK IKIIAPPERK YSVWIGGSIL ASLSTFQQMW ISKQEYDESG PSIVHRKCF UniProtKB: Actin, cytoplasmic 1 |
-Macromolecule #5: Isoform 3 of Gamma-tubulin complex component 2
| Macromolecule | Name: Isoform 3 of Gamma-tubulin complex component 2 / type: protein_or_peptide / ID: 5 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 105.5815 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSEFRIHHDV NELLSLLRVH GGDGAEVYID LLQKNRTPYV TTTVSAHSAK VKIAEFSRTP EDFLKKYDEL KSKNTRNLDP LVYLLSKLT EDKETLQYLQ QNAKERAELA AAAVGSSTTS INVPAAASKI SMQELEELRK QLGSVATGST LQQSLELKRK M LRDKQNKK ...String: MSEFRIHHDV NELLSLLRVH GGDGAEVYID LLQKNRTPYV TTTVSAHSAK VKIAEFSRTP EDFLKKYDEL KSKNTRNLDP LVYLLSKLT EDKETLQYLQ QNAKERAELA AAAVGSSTTS INVPAAASKI SMQELEELRK QLGSVATGST LQQSLELKRK M LRDKQNKK NSGQHLPIFP AWVYERPALI GDFLIGAGIS TDTALPIVLL GWSLALSPRL KCSGVISAHC GLHLPGTLPL AS QESAVVE DLLYVLVGVD GRYVSAQPLA GRQSRTFLVD PNLDLSIREL VHRILPVAAS YSAVTRFIEE KSSFEYGQVN HAL AAAMRT LVKEHLILVS QLEQLHRQGL LSLQKLWFYI QPAMRTMDIL ASLATSVDKG ECLGGSTLSL LHDRSFSYTG DSQA QELCL YLTKAASAPY FEVLEKWIYR GIIHDPYSEF MVEEHELRKE RIQEDYNDKY WDQRYTIVQQ QIPSFLQKMA DKILS TGKY LNVVRECGHD VTCPVAKEII YTLKERAYVE QIEKAFNYAS KVLLDFLMEE KELVAHLRSI KRYFLMDQGD FFVHFM DLA EEELRKPVED ITPPRLEALL ELALRMSTAN TDPFKDDLKI DLMPHDLITQ LLRVLAIETK QEKAMAHADP TELALSG LE AFSFDYIVKW PLSLIINRKA LTRYQMLFRH MFYCKHVERQ LCSVWISNKT AKQHSLHSAQ WFAGAFTLRQ RMLNFVQN I QYYMMFEVME PTWHILEKNL KSASNIDDVL GHHTGFLDTC LKDCMLTNPE LLKVFSKLMS VCVMFTNCMQ KFTQSMKLD GELGGQTLEH STVLGLPAGA EERARKELAR KHLAEHADTV QLVSGFEATI NKFDKNFSAH LLDLLARLSI YSTSDCEHGM ASVISRLDF NGFYTERLER LSAERSQKAT PQVPVLRGPP APAPRVAVTA Q UniProtKB: Gamma-tubulin complex component 2 |
-Macromolecule #6: Tubulin gamma-1 chain
| Macromolecule | Name: Tubulin gamma-1 chain / type: protein_or_peptide / ID: 6 / Number of copies: 14 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.022617 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MPREIITLQL GQCGNQIGFE FWKQLCAEHG ISPEGIVEEF ATEGTDRKDV FFYQADDEHY IPRAVLLDLE PRVIHSILNS PYAKLYNPE NIYLSEHGGG AGNNWASGFS QGEKIHEDIF DIIDREADGS DSLEGFVLCH SIAGGTGSGL GSYLLERLND R YPKKLVQT ...String: MPREIITLQL GQCGNQIGFE FWKQLCAEHG ISPEGIVEEF ATEGTDRKDV FFYQADDEHY IPRAVLLDLE PRVIHSILNS PYAKLYNPE NIYLSEHGGG AGNNWASGFS QGEKIHEDIF DIIDREADGS DSLEGFVLCH SIAGGTGSGL GSYLLERLND R YPKKLVQT YSVFPNQDEM SDVVVQPYNS LLTLKRLTQN ADCVVVLDNT ALNRIATDRL HIQNPSFSQI NQLVSTIMSA ST TTLRYPG YMNNDLIGLI ASLIPTPRLH FLMTGYTPLT TDQSVASVRK TTVLDVMRRL LQPKNVMVST GRDRQTNHCY IAI LNIIQG EVDPTQVHKS LQRIRERKLA NFIPWGPASI QVALSRKSPY LPSAHRVSGL MMANHTSISS LFERTCRQYD KLRK REAFL EQFRKEDMFK DNFDEMDTSR EIVQQLIDEY HAATRPDYIS WGTQEQENLY FQ UniProtKB: Tubulin gamma-1 chain |
-Macromolecule #7: Isoform 2 of Gamma-tubulin complex component 4
| Macromolecule | Name: Isoform 2 of Gamma-tubulin complex component 4 / type: protein_or_peptide / ID: 7 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 76.108898 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MIHELLLALS GYPGSIFTWN KRSGLQVSQD FPFLHPSETS VLNRLCRLGT DYIRFTEFIE QYTGHVQQQD HHPSQQGQGG LHGIYLRAF CTGLDSVLQP YRQALLDLEQ EFLGDPHLSI SHVNYFLDQF QLLFPSVMVV VEQIKSQKIH GCQILETVYK H SCGGLPPV ...String: MIHELLLALS GYPGSIFTWN KRSGLQVSQD FPFLHPSETS VLNRLCRLGT DYIRFTEFIE QYTGHVQQQD HHPSQQGQGG LHGIYLRAF CTGLDSVLQP YRQALLDLEQ EFLGDPHLSI SHVNYFLDQF QLLFPSVMVV VEQIKSQKIH GCQILETVYK H SCGGLPPV RSALEKILAV CHGVMYKQLS AWMLHGLLLD QHEEFFIKQG PSSGNVSAQP EEDEEDLGIG GLTGKQLREL QD LRLIEEE NMLAPSLKQF SLRVEILPSY IPVRVAEKIL FVGESVQMFE NQNVNLTRKG SILKNQEDTF AAELHRLKQQ PLF SLVDFE QVVDRIRSTV AEHLWKLMVE ESDLLGQLKI IKDFYLLGRG ELFQAFIDTA QHMLKTPPTA VTEHDVNVAF QQSA HKVLL DDDNLLPLLH LTIEYHGKEH KDATQAREGP SRETSPREAP ASGWAALGLS YKVQWPLHIL FTPAVLEKYN VVFKY LLSV RRVQAELQHC WALQMQRKHL KSNQTDAIKW RLRNHMAFLV DNLQYYLQVD VLESQFSQLL HQINSTRDFE SIRLAH DHF LSNLLAQSFI LLKPVFHCLN EILDLCHSFC SLVSQNLGPL DERGAAQLSI LVKGFSRQSS LLFKILSSVR NHQINSD LA QLLLRLDYNK YYTQAGGTLG SFGM UniProtKB: Gamma-tubulin complex component 4 |
-Macromolecule #8: Gamma-tubulin complex component 5
| Macromolecule | Name: Gamma-tubulin complex component 5 / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 118.367406 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MARHGPPWSR LDAQQERDVR ELVRGVAGLQ DEADPNFQLA LNFAWSNFRF HRFLDVNSHK IEKTIEGIYE KFVIHSDLSK AASWKRLTE EFLNAPLPSI KEIKTDAHYS ILSLLLCLSD SPSNSSYVET PRNKEVEKKD DFDWGKYLME DEEMDIGPYM D TPNWSEES ...String: MARHGPPWSR LDAQQERDVR ELVRGVAGLQ DEADPNFQLA LNFAWSNFRF HRFLDVNSHK IEKTIEGIYE KFVIHSDLSK AASWKRLTE EFLNAPLPSI KEIKTDAHYS ILSLLLCLSD SPSNSSYVET PRNKEVEKKD DFDWGKYLME DEEMDIGPYM D TPNWSEES EEENDQQPLS REDSGIQVDR TPLEEQDQNG KLDPCISWKD EPDDRSWLEH HVVHQYWTAR PSQFPHSLHL HS NLAAVWD QHLYSSDPLY VPDDRVLVTE TQVIRETLWL LSGVKKLFIF QLIDGKVTVR NNIIVTHLTH SCLRSVLEQI AAY GQVVFR LQEFIDEVMG HSSESMLPGS GSVPKKSTEA PFRTYQAFMW ALYKYFISFK EELAEIEKCI INNDTTITLA IVVD KLAPR LSQLKVLHKV FSTGVAEVPP DTRNVVRASH LLNTLYKAIL EYDNVGEASE QTVSLLFSLW VETVRPYLQT VDEWI VHGH LWDGAREFII QRNKNVPVNH RDFWYATYTL YSVSEKTENE EKMSDNASAS SGSDQGPSSR QHTMVSFLKP VLKQII MAG KSMQLLKNLQ CAESTTCQAG ARDAERKSLY TLFLESVQSR LRHGEDSTPQ VLTEQQATKE NLMKMQSIAE SHLELDD VH DPLLAINFAR MYLEQSDFHE KFAGGDVCVD RSSESVTCQT FELTLRSCLY PHIDKQYLDC CGNLMQTLKK DYRLVEYL Q AMRNFFLMEG GDTMYDFYTS IFDKIREKET WQNVSFLNVQ LQEAVGQRYP EDSSRLSISF ENVDTAKKKL PVHILDGLT LSYKVPWPVD IVISLECQKI YNQVFLLLLQ IKWAKYSLDV LLFGELVSTA EKPRLKEGLI HEQDTVAQFG PQKEPVRQQI HRMFLLRVK LMHFVNSLHN YIMTRILHST GLEFQHQVEE AKDLDQLIKI HYRYLSTIHD RCLLREKVSF VKEAIMKVLN L ALMFADGW QAGLGTWRME SIEKMESDFK NCHMFLVTIL NKAVCRGSFP HLESLALSLM AGMEQS UniProtKB: Gamma-tubulin complex component 5 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8vrd: |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)