+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CCP5 in complex with microtubules class1 | |||||||||
 Map data Map data | Composite map of CCP5 class1 in complex with two microtubule protofilaments, stitched from deepEMhancer processed focused CCP5 map and two protofilament maps. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | carboxypeptidase deglutamylation branch glutamate removal microtubule / HYDROLASE / HYDROLASE-SUBSTRATE complex / HYDROLASE-SUBSTRATE / STRUCTURAL PROTEIN complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationtubulin-glutamate carboxypeptidase / protein deglutamylation / protein side chain deglutamylation / protein branching point deglutamylation / C-terminal protein deglutamylation / Carboxyterminal post-translational modifications of tubulin / Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Resolution of Sister Chromatid Cohesion / Hedgehog 'off' state / Cilium Assembly ...tubulin-glutamate carboxypeptidase / protein deglutamylation / protein side chain deglutamylation / protein branching point deglutamylation / C-terminal protein deglutamylation / Carboxyterminal post-translational modifications of tubulin / Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Resolution of Sister Chromatid Cohesion / Hedgehog 'off' state / Cilium Assembly / Intraflagellar transport / COPI-dependent Golgi-to-ER retrograde traffic / Mitotic Prometaphase / Carboxyterminal post-translational modifications of tubulin / RHOH GTPase cycle / EML4 and NUDC in mitotic spindle formation / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / PKR-mediated signaling / Separation of Sister Chromatids / The role of GTSE1 in G2/M progression after G2 checkpoint / Aggrephagy / RHO GTPases activate IQGAPs / RHO GTPases Activate Formins / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / MHC class II antigen presentation / Recruitment of NuMA to mitotic centrosomes / COPI-mediated anterograde transport / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / metallocarboxypeptidase activity / tubulin binding / structural constituent of cytoskeleton / microtubule cytoskeleton organization / neuron migration / mitotic spindle / mitotic cell cycle / microtubule cytoskeleton / midbody / defense response to virus / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / GTPase activity / GTP binding / proteolysis / zinc ion binding / metal ion binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Chen J / Zehr EA / Gruschus JM / Szyk A / Liu Y / Tanner ME / Tjandra N / Roll-Mecak A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Tubulin code eraser CCP5 binds branch glutamates by substrate deformation. Authors: Jiayi Chen / Elena A Zehr / James M Gruschus / Agnieszka Szyk / Yanjie Liu / Martin E Tanner / Nico Tjandra / Antonina Roll-Mecak /   Abstract: Microtubule function is modulated by the tubulin code, diverse posttranslational modifications that are altered dynamically by writer and eraser enzymes. Glutamylation-the addition of branched ...Microtubule function is modulated by the tubulin code, diverse posttranslational modifications that are altered dynamically by writer and eraser enzymes. Glutamylation-the addition of branched (isopeptide-linked) glutamate chains-is the most evolutionarily widespread tubulin modification. It is introduced by tubulin tyrosine ligase-like enzymes and erased by carboxypeptidases of the cytosolic carboxypeptidase (CCP) family. Glutamylation homeostasis, achieved through the balance of writers and erasers, is critical for normal cell function, and mutations in CCPs lead to human disease. Here we report cryo-electron microscopy structures of the glutamylation eraser CCP5 in complex with the microtubule, and X-ray structures in complex with transition-state analogues. Combined with NMR analysis, these analyses show that CCP5 deforms the tubulin main chain into a unique turn that enables lock-and-key recognition of the branch glutamate in a cationic pocket that is unique to CCP family proteins. CCP5 binding of the sequences flanking the branch point primarily through peptide backbone atoms enables processing of diverse tubulin isotypes and non-tubulin substrates. Unexpectedly, CCP5 exhibits inefficient processing of an abundant β-tubulin isotype in the brain. This work provides an atomistic view into glutamate branch recognition and resolution, and sheds light on homeostasis of the tubulin glutamylation syntax. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42971.map.gz emd_42971.map.gz | 252.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42971-v30.xml emd-42971-v30.xml emd-42971.xml emd-42971.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42971.png emd_42971.png | 165 KB | ||
| Filedesc metadata |  emd-42971.cif.gz emd-42971.cif.gz | 7.6 KB | ||
| Others |  emd_42971_additional_1.map.gz emd_42971_additional_1.map.gz | 279.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42971 http://ftp.pdbj.org/pub/emdb/structures/EMD-42971 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42971 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42971 | HTTPS FTP |
-Related structure data
| Related structure data |  8v4kMC  8v3mC  8v3nC  8v3oC  8v3pC  8v3qC  8v3rC  8v3sC  8v4lC  8v4mC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42971.map.gz / Format: CCP4 / Size: 381.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42971.map.gz / Format: CCP4 / Size: 381.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map of CCP5 class1 in complex with two microtubule protofilaments, stitched from deepEMhancer processed focused CCP5 map and two protofilament maps. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.245 Å | ||||||||||||||||||||||||||||||||||||
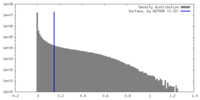
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Composite raw full map of CCP5 class1 in...
| File | emd_42971_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite raw full map of CCP5 class1 in complex with two microtubule protofilaments, stitched from CCP5 focused and two protofilament raw full maps. | ||||||||||||
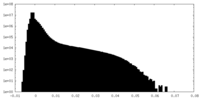
| Projections & Slices |
| ||||||||||||
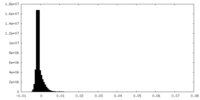
| Density Histograms |
- Sample components
Sample components
-Entire : CCP5 in complex with microtubules
| Entire | Name: CCP5 in complex with microtubules |
|---|---|
| Components |
|
-Supramolecule #1: CCP5 in complex with microtubules
| Supramolecule | Name: CCP5 in complex with microtubules / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1, #3 / Details: Composite structure of CCP5:microtubule class#1 |
|---|
-Macromolecule #1: Tubulin alpha-1B chain
| Macromolecule | Name: Tubulin alpha-1B chain / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.204445 KDa |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE FSIYPAPQVS TAVVEPYNSI LTTHTTLEHS DCAFMVDNEA IYDICRRNLD IERPTYTNLN RLISQIVSSI TA SLRFDGA LNVDLTEFQT NLVPYPRIHF PLATYAPVIS AEKAYHEQLS VAEITNACFE PANQMVKCDP RHGKYMACCL LYR GDVVPK DVNAAIATIK TKRSIQFVDW CPTGFKVGIN YQPPTVVPGG DLAKVQRAVC MLSNTTAIAE AWARLDHKFD LMYA KRAFV HWYVGEGMEE GEFSEAREDM AALEKDYEEV GVDSVEGEGE EEGEEY UniProtKB: Tubulin alpha-1B chain |
-Macromolecule #2: Tubulin beta chain
| Macromolecule | Name: Tubulin beta chain / type: protein_or_peptide / ID: 2 Details: Beta tubulin from porcine brain microtubules, heterogeneous in tail sequence Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.504801 KDa |
| Sequence | String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS ...String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS VVPSPKVSDT VVEPYNATLS VHQLVENTDE TYCIDNEALY DICFRTLKLT TPTYGDLNHL VSATMSGVTT CL RFPGQLN ADLRKLAVNM VPFPRLHFFM PGFAPLTSRG SQQYRALTVP ELTQQMFDAK NMMAACDPRH GRYLTVAAVF RGR MSMKEV DEQMLNVQNK NSSYFVEWIP NNVKTAVCDI PPRGLKMSAT FIGNSTAIQE LFKRISEQFT AMFRRKAFLH WYTG EGMDE MEFTEAESNM NDLVSEYQQY QDATA(UNK)(UNK)(UNK)(UNK)E (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK) UniProtKB: Tubulin beta chain |
-Macromolecule #3: Cytosolic carboxypeptidase-like protein 5
| Macromolecule | Name: Cytosolic carboxypeptidase-like protein 5 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 68.229547 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NELRCGGLLF SSRFDSGNLA HVEKVESLSS DGEGVGGGAS ALTSGIASSP DYEFNVWTRP DCAETEFENG NRSWFYFSVR GGMPGKLIK INIMNMNKQS KLYSQGMAPF VRTLPTRPRW ERIRDRPTFE MTETQFVLSF VHRFVEGRGA TTFFAFCYPF S YSDCQELL ...String: NELRCGGLLF SSRFDSGNLA HVEKVESLSS DGEGVGGGAS ALTSGIASSP DYEFNVWTRP DCAETEFENG NRSWFYFSVR GGMPGKLIK INIMNMNKQS KLYSQGMAPF VRTLPTRPRW ERIRDRPTFE MTETQFVLSF VHRFVEGRGA TTFFAFCYPF S YSDCQELL NQLDQRFPEN HPTHSSPLDT IYYHRELLCY SLDGLRVDLL TITSCHGLRE DREPRLEQLF PDTSTPRPFR FA GKRIFFL SSRVHPGETP SSFVFNGFLD FILRPDDPRA QTLRRLFVFK LIPMLNPDGV VRGHYRTDSR GVNLNRQYLK PDA VLHPAI YGAKAVLLYH HVHSRLNSQS SSEHQPSSCL PPDAPVSDLE KANNLQNEAQ CGHSADRHNA EAWKQTEPAE QKLN SVWIM PQQSAGLEES APDTIPPKES GVAYYVDLHG HASKRGCFMY GNSFSDESTQ VENMLYPKLI SLNSAHFDFQ GCNFS EKNM YARDRRDGQS KEGSGRVAIY KASGIIHSYT LACNYNTGRS VNSIPAACHD NGRASPPPPP AFPSRYTVEL FEQVGR AMA IAALDMAECN PWPRIVLSEH SSLTNLRAWM LKHVRNSRGL SS UniProtKB: Cytosolic carboxypeptidase-like protein 5 |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Macromolecule #6: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER
| Macromolecule | Name: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER / type: ligand / ID: 6 / Number of copies: 2 / Formula: G2P |
|---|---|
| Molecular weight | Theoretical: 521.208 Da |
| Chemical component information |  ChemComp-G2P: |
-Macromolecule #7: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 7 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #8: GLUTAMIC ACID
| Macromolecule | Name: GLUTAMIC ACID / type: ligand / ID: 8 Details: enzymatically linked to the side chain of GLU B 435 of beta-tubulin Number of copies: 1 / Formula: GLU |
|---|---|
| Molecular weight | Theoretical: 147.129 Da |
| Chemical component information |  ChemComp-GLU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Au-flat 1.2/1.3 / Material: GOLD / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 45 | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 303 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 1.651 sec. / Average electron dose: 53.34 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 135000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
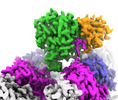
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation coefficient | |||||||||
| Output model |  PDB-8v4k: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)