[English] 日本語
 Yorodumi
Yorodumi- EMDB-42054: Structure of Semliki Forest virus VLP in complex with VLDLR LA2 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Semliki Forest virus VLP in complex with VLDLR LA2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Semliki Forest Virus / Receptor / VIRUS LIKE PARTICLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationreelin receptor activity / VLDL clearance / glycoprotein transport / very-low-density lipoprotein particle receptor activity / Reelin signalling pathway / ventral spinal cord development / very-low-density lipoprotein particle binding / low-density lipoprotein particle receptor activity / very-low-density lipoprotein particle clearance / togavirin ...reelin receptor activity / VLDL clearance / glycoprotein transport / very-low-density lipoprotein particle receptor activity / Reelin signalling pathway / ventral spinal cord development / very-low-density lipoprotein particle binding / low-density lipoprotein particle receptor activity / very-low-density lipoprotein particle clearance / togavirin / reelin-mediated signaling pathway / very-low-density lipoprotein particle / positive regulation of dendrite development / cargo receptor activity / T=4 icosahedral viral capsid / lipid transport / virion assembly / dendrite morphogenesis / regulation of synapse assembly / small molecule binding / apolipoprotein binding / cholesterol metabolic process / clathrin-coated pit / receptor-mediated endocytosis / VLDLR internalisation and degradation / memory / calcium-dependent protein binding / nervous system development / clathrin-dependent endocytosis of virus by host cell / host cell endosome / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / receptor complex / serine-type endopeptidase activity / viral translational frameshifting / lysosomal membrane / fusion of virus membrane with host endosome membrane / viral envelope / calcium ion binding / symbiont entry into host cell / virion attachment to host cell / host cell nucleus / host cell plasma membrane / virion membrane / glutamatergic synapse / structural molecule activity / signal transduction / proteolysis / RNA binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Semliki Forest virus / Semliki Forest virus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Abraham J / Yang P / Li W / Fan X / Pan J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for VLDLR recognition by eastern equine encephalitis virus. Authors: Pan Yang / Wanyu Li / Xiaoyi Fan / Junhua Pan / Colin J Mann / Haley Varnum / Lars E Clark / Sarah A Clark / Adrian Coscia / Himanish Basu / Katherine Nabel Smith / Vesna Brusic / Jonathan Abraham /   Abstract: Eastern equine encephalitis virus (EEEV) is the most virulent alphavirus that infects humans, and many survivors develop neurological sequelae, including paralysis and intellectual disability. ...Eastern equine encephalitis virus (EEEV) is the most virulent alphavirus that infects humans, and many survivors develop neurological sequelae, including paralysis and intellectual disability. Alphavirus spike proteins comprise trimers of heterodimers of glycoproteins E2 and E1 that mediate binding to cellular receptors and fusion of virus and host cell membranes during entry. We recently identified very-low density lipoprotein receptor (VLDLR) and apolipoprotein E receptor 2 (ApoER2) as cellular receptors for EEEV and a distantly related alphavirus, Semliki Forest virus (SFV). Here, we use single-particle cryo-electron microscopy (cryo-EM) to determine structures of the EEEV and SFV spike glycoproteins bound to the VLDLR ligand-binding domain and found that EEEV and SFV interact with the same cellular receptor through divergent binding modes. Our studies suggest that the ability of LDLR-related proteins to interact with viral spike proteins through very small footprints with flexible binding modes results in a low evolutionary barrier to the acquisition of LDLR-related proteins as cellular receptors for diverse sets of viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42054.map.gz emd_42054.map.gz | 272.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42054-v30.xml emd-42054-v30.xml emd-42054.xml emd-42054.xml | 25.2 KB 25.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42054.png emd_42054.png | 61.9 KB | ||
| Filedesc metadata |  emd-42054.cif.gz emd-42054.cif.gz | 7.2 KB | ||
| Others |  emd_42054_additional_1.map.gz emd_42054_additional_1.map.gz emd_42054_half_map_1.map.gz emd_42054_half_map_1.map.gz emd_42054_half_map_2.map.gz emd_42054_half_map_2.map.gz | 275.7 MB 241.6 MB 241.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42054 http://ftp.pdbj.org/pub/emdb/structures/EMD-42054 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42054 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42054 | HTTPS FTP |
-Related structure data
| Related structure data |  8ua8MC  8ua4C  8ua9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42054.map.gz / Format: CCP4 / Size: 303.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42054.map.gz / Format: CCP4 / Size: 303.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
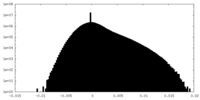
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_42054_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
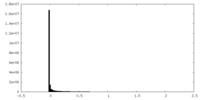
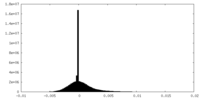
| Density Histograms |
-Half map: #2
| File | emd_42054_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
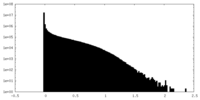
| Density Histograms |
-Half map: #1
| File | emd_42054_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
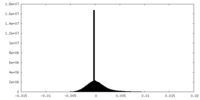
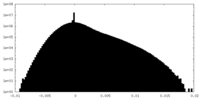
| Density Histograms |
- Sample components
Sample components
-Entire : Semliki Forest virus
| Entire | Name:   Semliki Forest virus Semliki Forest virus |
|---|---|
| Components |
|
-Supramolecule #1: Semliki Forest virus
| Supramolecule | Name: Semliki Forest virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 / NCBI-ID: 11033 / Sci species name: Semliki Forest virus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: Glycoprotein E1
| Macromolecule | Name: Glycoprotein E1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Semliki Forest virus Semliki Forest virus |
| Molecular weight | Theoretical: 47.489766 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: YEHSTVMPNV VGFPYKAHIE RPGYSPLTLQ MQVVETSLEP TLNLEYITCE YKTVVPSPYV KCCGASECST KEKPDYQCKV YTGVYPFMW GGAYCFCDSE NTQLSEAYVD RSDVCRHDHA SAYKAHTASL KAKVRVMYGN VNQTVDVYVN GDHAVTIGGT Q FIFGPLSS ...String: YEHSTVMPNV VGFPYKAHIE RPGYSPLTLQ MQVVETSLEP TLNLEYITCE YKTVVPSPYV KCCGASECST KEKPDYQCKV YTGVYPFMW GGAYCFCDSE NTQLSEAYVD RSDVCRHDHA SAYKAHTASL KAKVRVMYGN VNQTVDVYVN GDHAVTIGGT Q FIFGPLSS AWTPFDNKIV VYKDEVFNQD FPPYGSGQPG RFGDIQSRTV ESNDLYANTA LKLARPSPGM VHVPYTQTPS GF KYWLKEK GTALNTKAPF GCQIKTNPVR AMNCAVGNIP VSMNLPDSAF TRIVEAPTII DLTCTVATCT HSSDFGGVLT LTY KTDKNG DCSVHSHSNV ATLQEATAKV KTAGKVTLHF STASASPSFV VSLCSARATC SASCEPPKDH IVPYAASHSN VVFP DMSGT ALSWVQKISG GLGAFAIGAI LVLVVVTCIG LRR UniProtKB: Structural polyprotein |
-Macromolecule #2: Glycoprotein E2
| Macromolecule | Name: Glycoprotein E2 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Semliki Forest virus Semliki Forest virus |
| Molecular weight | Theoretical: 46.330719 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: FNVYKATRPY IAYCADCGAG HSCHSPVAIE AVRSEATDGM LKIQFSAQIG IDKSDNHDYT KIRYADGHAI ENAVRSSLKV ATSGDCFVH GTMGHFILAK CPPGEFLQVS IQDTRNAVRA CRIQYHHDPQ PVGREKFTIR PHYGKEIPCT TYQQTTAKTV E EIDMHMPP ...String: FNVYKATRPY IAYCADCGAG HSCHSPVAIE AVRSEATDGM LKIQFSAQIG IDKSDNHDYT KIRYADGHAI ENAVRSSLKV ATSGDCFVH GTMGHFILAK CPPGEFLQVS IQDTRNAVRA CRIQYHHDPQ PVGREKFTIR PHYGKEIPCT TYQQTTAKTV E EIDMHMPP DTPDRTLLSQ QSGNVKITVG GKKVKYNCTC GTGNVGTTNS DMTINTCLIE QCHVSVTDHK KWQFNSPFVP RA DEPARKG KVHIPFPLDN ITCRVPMARE PTVIHGKREV TLHLHPDHPT LFSYRTLGED PQYHEEWVTA AVERTIPVPV DGM EYHWGN NDPVRLWSQL TTEGKPHGWP HQIVQYYYGL YPAATVSAVV GMSLLALISI FASCYMLVAA RSKCLTPYAL TPGA AVPWT LGILCCAPRA HA UniProtKB: UNIPROTKB: A0A0E3T652 |
-Macromolecule #3: Assembly protein E3
| Macromolecule | Name: Assembly protein E3 / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Semliki Forest virus Semliki Forest virus |
| Molecular weight | Theoretical: 6.020911 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TAMCVLANAT FPCFQPPCVP CCYENNAEAT LRMLEDNVDR PGYYDLLQAA LTCR UniProtKB: Structural polyprotein |
-Macromolecule #4: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 4 / Number of copies: 4 / Enantiomer: LEVO / EC number: togavirin |
|---|---|
| Source (natural) | Organism:   Semliki Forest virus Semliki Forest virus |
| Molecular weight | Theoretical: 16.723904 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: IENDCIFEVK HEGKVTGYAC LVGDKVMKPA HVKGVIDNAD LAKLAFKKSS KYDLECAQIP VHMRSDASKY THEKPEGHYN WHHGAVQYS GGRFTIPTGA GKPGDSGRPI FDNKGRVVAI VLGGANEGSR TALSVVTWNK DMVTRVTPEG SEEW UniProtKB: Structural polyprotein |
-Macromolecule #5: Very low-density lipoprotein receptor
| Macromolecule | Name: Very low-density lipoprotein receptor / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 4.028225 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: CAESDFVCNN GQCVPSRWKC DGDPDCEDGS DESPEQC UniProtKB: Very low-density lipoprotein receptor |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #9: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 9 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 54.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)