[English] 日本語
 Yorodumi
Yorodumi- EMDB-4190: A mechanism for the activation of the influenza virus transcriptase -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4190 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | A mechanism for the activation of the influenza virus transcriptase | ||||||||||||
 Map data Map data | EM map of influenza C virus RNA dependent RNA polymerase bound to promoter vRNA | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | influenza virus RNA dependent RNA polymerase / VIRAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationcap snatching / viral transcription / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / host cell mitochondrion / 7-methylguanosine mRNA capping / virion component / endonuclease activity / Hydrolases; Acting on ester bonds / host cell cytoplasm / symbiont-mediated suppression of host gene expression ...cap snatching / viral transcription / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / host cell mitochondrion / 7-methylguanosine mRNA capping / virion component / endonuclease activity / Hydrolases; Acting on ester bonds / host cell cytoplasm / symbiont-mediated suppression of host gene expression / viral translational frameshifting / hydrolase activity / RNA-directed RNA polymerase / viral RNA genome replication / nucleotide binding / RNA-directed RNA polymerase activity / DNA-templated transcription / host cell nucleus / RNA binding / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Influenza B virus / Influenza B virus /  Influenza B virus (B/Memphis/13/2003) Influenza B virus (B/Memphis/13/2003) | ||||||||||||
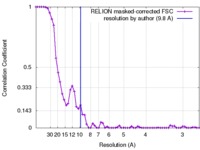
| Method | single particle reconstruction / cryo EM / Resolution: 9.8 Å | ||||||||||||
 Authors Authors | Serna Martin I / Grimes JM | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2018 Journal: Mol Cell / Year: 2018Title: A Mechanism for the Activation of the Influenza Virus Transcriptase. Authors: Itziar Serna Martin / Narin Hengrung / Max Renner / Jane Sharps / Mónica Martínez-Alonso / Simonas Masiulis / Jonathan M Grimes / Ervin Fodor /  Abstract: Influenza virus RNA polymerase (FluPol), a heterotrimer composed of PB1, PB2, and PA subunits (P3 in influenza C), performs both transcription and replication of the viral RNA genome. For ...Influenza virus RNA polymerase (FluPol), a heterotrimer composed of PB1, PB2, and PA subunits (P3 in influenza C), performs both transcription and replication of the viral RNA genome. For transcription, FluPol interacts with the C-terminal domain (CTD) of RNA polymerase II (Pol II), which enables FluPol to snatch capped RNA primers from nascent host RNAs. Here, we describe the co-crystal structure of influenza C virus polymerase (FluPol) bound to a Ser5-phosphorylated CTD (pS-CTD) peptide. The position of the CTD-binding site at the interface of PB1, P3, and the flexible PB2 C-terminal domains suggests that CTD binding stabilizes the transcription-competent conformation of FluPol. In agreement, both cap snatching and capped primer-dependent transcription initiation by FluPol are enhanced in the presence of pS-CTD. Mutations of amino acids in the CTD-binding site reduce viral mRNA synthesis. We propose a model for the activation of the influenza virus transcriptase through its association with pS-CTD of Pol II. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4190.map.gz emd_4190.map.gz | 2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4190-v30.xml emd-4190-v30.xml emd-4190.xml emd-4190.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4190_fsc.xml emd_4190_fsc.xml | 5.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_4190.png emd_4190.png | 57 KB | ||
| Filedesc metadata |  emd-4190.cif.gz emd-4190.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4190 http://ftp.pdbj.org/pub/emdb/structures/EMD-4190 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4190 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4190 | HTTPS FTP |
-Related structure data
| Related structure data |  6f5oMC  6f5pC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4190.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4190.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map of influenza C virus RNA dependent RNA polymerase bound to promoter vRNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Influenza C virus polymerase bound to promoter vRNA
| Entire | Name: Influenza C virus polymerase bound to promoter vRNA |
|---|---|
| Components |
|
-Supramolecule #1: Influenza C virus polymerase bound to promoter vRNA
| Supramolecule | Name: Influenza C virus polymerase bound to promoter vRNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Influenza B virus Influenza B virus |
-Macromolecule #1: Polymerase acidic protein
| Macromolecule | Name: Polymerase acidic protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza B virus Influenza B virus |
| Molecular weight | Theoretical: 83.232109 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SMDTFITRNF QTTIIQKAKN TMAEFSEDPE LQPAMLFNIC VHLEVCYVIS DMNFLDEEGK AYTALEGQGK EQNLRPQYEV IEGMPRTIA WMVQRSLAQE HGIETPKYLA DLFDYKTKRF IEVGITKGLA DDYFWKKKEK LGNSMELMIF SYNQDYSLSN E SSLDEEGK ...String: SMDTFITRNF QTTIIQKAKN TMAEFSEDPE LQPAMLFNIC VHLEVCYVIS DMNFLDEEGK AYTALEGQGK EQNLRPQYEV IEGMPRTIA WMVQRSLAQE HGIETPKYLA DLFDYKTKRF IEVGITKGLA DDYFWKKKEK LGNSMELMIF SYNQDYSLSN E SSLDEEGK GRVLSRLTEL QAELSLKNLW QVLIGEEDVE KGIDFKLGQT ISRLRDISVP AGFSNFEGMR SYIDNIDPKG AI ERNLARM SPLVSVTPKK LTWEDLRPIG PHIYNHELPE VPYNAFLLMS DELGLANMTE GKSKKPKTLA KECLEKYSTL RDQ TDPILI MKSEKANENF LWKLWRDCVN TISNEEMSNE LQKTNYAKWA TGDGLTYQKI MKEVAIDDET MCQEEPKIPN KCRV AAWVQ TEMNLLSTLT SKRALDLPEI GPDVAPVEHV GSERRKYFVN EINYCKASTV MMKYVLFHTS LLNESNASMG KYKVI PITN RVVNEKGESF DMLYGLAVKG QSHLRGDTDV VTVVTFEFSS TDPRVDSGKW PKYTVFRIGS LFVSGREKSV YLYCRV NGT NKIQMKWGME ARRCLLQSMQ QMEAIVEQES SIQGYDMTKA CFKGDRVNSP KTFSIGTQEG KLVKGSFGKA LRVIFTK CL MHYVFGNAQL EGFSAESRRL LLLIQALKDR KGPWVFDLEG MYSGIEECIS NNPWVIQSAY WFNEWLGFEK EGSKVLES V DEIMDE UniProtKB: Polymerase acidic protein |
-Macromolecule #2: RNA-directed RNA polymerase catalytic subunit
| Macromolecule | Name: RNA-directed RNA polymerase catalytic subunit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Influenza B virus Influenza B virus |
| Molecular weight | Theoretical: 84.433266 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNINPYFLFI DVPIQAAIST TFPYTGVPPY SHGTGTGYTI DTVIRTHEYS NKGKQYISDV TGCTMVDPTN GPLPEDNEPS AYAQLDCVL EALDRMDEEH PGLFQAASQN AMETLMVTTV DKLTQGRQTF DWTVCRNQPA ATALNTTITS FRLNDLNGAD K GGLIPFCQ ...String: MNINPYFLFI DVPIQAAIST TFPYTGVPPY SHGTGTGYTI DTVIRTHEYS NKGKQYISDV TGCTMVDPTN GPLPEDNEPS AYAQLDCVL EALDRMDEEH PGLFQAASQN AMETLMVTTV DKLTQGRQTF DWTVCRNQPA ATALNTTITS FRLNDLNGAD K GGLIPFCQ DIIDSLDRPE MTFFSVKNIK KKLPAKNRKG FLIKRIPMKV KDKITKVEYI KRALSLNTMT KDAERGKLKR RA IATAGIQ IRGFVLVVEN LAKNICENLE QSGLPVGGNE KKAKLSNAVA KMLSNCPPGG ISMTVTGDNT KWNECLNPRI FLA MTERIT RDSPIWFRDF CSIAPVLFSN KIARLGKGFM ITSKTKRLKA QIPCPDLFSI PLERYNEETR AKLKKLKPFF NEEG TASLS PGMMMGMFNM LSTVLGVAAL GIKNIGNKEY LWDGLQSSDD FALFVNAKDE ETCMEGINDF YRTCKLLGIN MSKKK SYCN ETGMFEFTSM FYRDGFVSNF AMELPSFGVA GVNESADMAI GMTIIKNNMI NNGMGPATAQ TAIQLFIADY RYTYKC HRG DSKVEGKRMK IIKELWENTK GRDGLLVADG GPNIYNLRNL HIPEIVLKYN LMDPEYKGRL LHPQNPFVGH LSIEGIK EA DITPAHGPVK KMDYDAVSGT HSWRTKRNRS ILNTDQRNMI LEEQCYAKCC NLFEACFNSA SYRKPVGQHS MLEAMAHR L RMDARLDYES GRMSKDDFEK AMAHLGEIGY I UniProtKB: RNA-directed RNA polymerase catalytic subunit |
-Macromolecule #3: Polymerase basic protein 2
| Macromolecule | Name: Polymerase basic protein 2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza B virus Influenza B virus |
| Molecular weight | Theoretical: 88.028211 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTLAKIELLK QLLRDNEAKT VLKQTTVDQY NIIRKFNTSR IEKNPSLRMK WAMCSNFPLA LTKGDMANRI PLEYKGIQLK TNAEDIGTK GQMCSIAAVT WWNTYGPIGD TEGFERVYES FFLRKMRLDN ATWGRITFGP VERVRKRVLL NPLTKEMPPD E ASNVIMEI ...String: MTLAKIELLK QLLRDNEAKT VLKQTTVDQY NIIRKFNTSR IEKNPSLRMK WAMCSNFPLA LTKGDMANRI PLEYKGIQLK TNAEDIGTK GQMCSIAAVT WWNTYGPIGD TEGFERVYES FFLRKMRLDN ATWGRITFGP VERVRKRVLL NPLTKEMPPD E ASNVIMEI LFPKEAGIPR ESTWIHRELI KEKREKLKGT MITPIVLAYM LERELVARRR FLPVAGATSA EFIEMLHCLQ GE NWRQIYH PGGNKLTESR SQSMIVACRK IIRRSIVASN PLELAVEIAN KTVIDTEPLK SCLAAIDGGD VACDIIRAAL GLK IRQRQR FGRLELKRIS GRGFKNDEEI LIGNGTIQKI GIWDGEEEFH VRCGECRGIL KKSKMKLEKL LINSAKKEDM RDLI ILCMV FSQDTRMFQG VRGEINFLNR AGQLLSPMYQ LQRYFLNRSN DLFDQWGYEE SPKASELHGI NESMNASDYT LKGVV VTRN VIDDFSSTET EKVSITKNLS LIKRTGEVIM GANDVSELES QAQLMITYDT PKMWEMGTTK ELVQNTYQWV LKNLVT LKA QFLLGKEDMF QWDAFEAFES IIPQKMAGQY SGFARAVLKQ MRDQEVMKTD QFIKLLPFCF SPPKLRSNGE PYQFLKL VL KGGGENFIEV RKGSPLFSYN PQTEVLTICG RMMSLKGKIE DEERNRSMGN AVLAGFLVSG KYDPDLGDFK TIEELEKL K PGEKANILLY QGKPVKVVKR KRYSALSNDI SQGIKRQRMT VESMGWALS UniProtKB: Polymerase basic protein 2 |
-Macromolecule #4: 3' promoter vRNA
| Macromolecule | Name: 3' promoter vRNA / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Influenza B virus (B/Memphis/13/2003) Influenza B virus (B/Memphis/13/2003) |
| Molecular weight | Theoretical: 4.321563 KDa |
| Sequence | String: UAUACCUCUG CUUC |
-Macromolecule #5: 5' promoter vRNA
| Macromolecule | Name: 5' promoter vRNA / type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Influenza B virus (B/Memphis/13/2003) Influenza B virus (B/Memphis/13/2003) |
| Molecular weight | Theoretical: 4.55782 KDa |
| Sequence | String: AGUAGUAACA AGAG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 2.09 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6f5o: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)