[English] 日本語
 Yorodumi
Yorodumi- EMDB-41885: Structure of the HER2/HER4/BTC Heterodimer Extracellular Domain -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the HER2/HER4/BTC Heterodimer Extracellular Domain | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Receptor Tyrosine Kinase / MEMBRANE PROTEIN / TRANSFERASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationestablishment of planar polarity involved in nephron morphogenesis / ERBB4 signaling pathway / ERBB4-ERBB4 signaling pathway / olfactory bulb interneuron differentiation / central nervous system morphogenesis / neuregulin receptor activity / cardiac muscle tissue regeneration / negative regulation of neuron migration / negative regulation of immature T cell proliferation in thymus / ERBB3:ERBB2 complex ...establishment of planar polarity involved in nephron morphogenesis / ERBB4 signaling pathway / ERBB4-ERBB4 signaling pathway / olfactory bulb interneuron differentiation / central nervous system morphogenesis / neuregulin receptor activity / cardiac muscle tissue regeneration / negative regulation of neuron migration / negative regulation of immature T cell proliferation in thymus / ERBB3:ERBB2 complex / ERBB2-ERBB4 signaling pathway / GRB7 events in ERBB2 signaling / immature T cell proliferation in thymus / RNA polymerase I core binding / mitochondrial fragmentation involved in apoptotic process / semaphorin receptor complex / mammary gland epithelial cell differentiation / PI3K events in ERBB4 signaling / negative regulation of epithelial cell apoptotic process / embryonic pattern specification / GABA receptor binding / transmembrane receptor protein tyrosine kinase activator activity / positive regulation of urine volume / regulation of microtubule-based process / ErbB-3 class receptor binding / positive regulation of protein localization to cell surface / motor neuron axon guidance / Sema4D induced cell migration and growth-cone collapse / epidermal growth factor receptor binding / epithelial cell apoptotic process / neural crest cell migration / Inhibition of Signaling by Overexpressed EGFR / epidermal growth factor receptor activity / EGFR interacts with phospholipase C-gamma / positive regulation of tyrosine phosphorylation of STAT protein / PLCG1 events in ERBB2 signaling / ERBB2-EGFR signaling pathway / ERBB2 Activates PTK6 Signaling / enzyme-linked receptor protein signaling pathway / neurotransmitter receptor localization to postsynaptic specialization membrane / Signaling by EGFR / neuromuscular junction development / positive regulation of Rho protein signal transduction / ERBB2-ERBB3 signaling pathway / Drug-mediated inhibition of ERBB2 signaling / Resistance of ERBB2 KD mutants to trastuzumab / Resistance of ERBB2 KD mutants to sapitinib / Resistance of ERBB2 KD mutants to tesevatinib / Resistance of ERBB2 KD mutants to neratinib / Resistance of ERBB2 KD mutants to osimertinib / Resistance of ERBB2 KD mutants to afatinib / Resistance of ERBB2 KD mutants to AEE788 / Resistance of ERBB2 KD mutants to lapatinib / Drug resistance in ERBB2 TMD/JMD mutants / positive regulation of transcription by RNA polymerase I / positive regulation of MAP kinase activity / ERBB2 Regulates Cell Motility / semaphorin-plexin signaling pathway / oligodendrocyte differentiation / positive regulation of cell division / Signaling by ERBB4 / Long-term potentiation / PI3K events in ERBB2 signaling / mammary gland alveolus development / positive regulation of protein targeting to membrane / Estrogen-dependent nuclear events downstream of ESR-membrane signaling / SHC1 events in ERBB4 signaling / cell fate commitment / regulation of angiogenesis / cell surface receptor signaling pathway via JAK-STAT / GAB1 signalosome / regulation of ERK1 and ERK2 cascade / Schwann cell development / Nuclear signaling by ERBB4 / coreceptor activity / positive regulation of cardiac muscle cell proliferation / synapse assembly / lactation / Signaling by ERBB2 / TFAP2 (AP-2) family regulates transcription of growth factors and their receptors / myelination / GRB2 events in EGFR signaling / SHC1 events in EGFR signaling / transmembrane receptor protein tyrosine kinase activity / positive regulation of mitotic nuclear division / Downregulation of ERBB4 signaling / GRB2 events in ERBB2 signaling / positive regulation of cell adhesion / Downregulation of ERBB2:ERBB3 signaling / SHC1 events in ERBB2 signaling / cell surface receptor protein tyrosine kinase signaling pathway / basal plasma membrane / regulation of cell migration / Constitutive Signaling by Overexpressed ERBB2 / cellular response to epidermal growth factor stimulus / peptidyl-tyrosine phosphorylation / positive regulation of translation / positive regulation of epithelial cell proliferation / positive regulation of receptor signaling pathway via JAK-STAT / positive regulation of cell differentiation Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
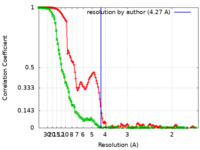
| Method | single particle reconstruction / cryo EM / Resolution: 4.27 Å | ||||||||||||
 Authors Authors | Trenker R / Diwanji D / Bingham T / Verba KA / Jura N | ||||||||||||
| Funding support |  Germany, Germany,  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation. Authors: Raphael Trenker / Devan Diwanji / Tanner Bingham / Kliment A Verba / Natalia Jura /  Abstract: Human Epidermal growth factor Receptor 4 (HER4 or ERBB4) carries out essential functions in the development and maintenance of the cardiovascular and nervous systems. HER4 activation is regulated by ...Human Epidermal growth factor Receptor 4 (HER4 or ERBB4) carries out essential functions in the development and maintenance of the cardiovascular and nervous systems. HER4 activation is regulated by a diverse group of extracellular ligands including the neuregulin (NRG) family and betacellulin (BTC), which promote HER4 homodimerization or heterodimerization with other HER receptors. Important cardiovascular functions of HER4 are exerted via heterodimerization with its close homolog and orphan receptor, HER2. To date structural insights into ligand-mediated HER4 activation have been limited to crystallographic studies of HER4 ectodomain homodimers in complex with NRG1β. Here, we report cryo-EM structures of near full-length HER2/HER4 heterodimers and full-length HER4 homodimers bound to NRG1β and BTC. We show that the structures of the heterodimers bound to either ligand are nearly identical and that in both cases the HER2/HER4 heterodimer interface is less dynamic than those observed in structures of HER2/EGFR and HER2/HER3 heterodimers. In contrast, structures of full-length HER4 homodimers bound to NRG1β and BTC display more large-scale dynamics mirroring states previously reported for EGFR homodimers. Our structures also reveal the presence of multiple glycan modifications within HER4 ectodomains, modeled for the first time in HER receptors, that distinctively contribute to the stabilization of HER4 homodimer interfaces over those of HER2/HER4 heterodimers. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41885.map.gz emd_41885.map.gz | 203.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41885-v30.xml emd-41885-v30.xml emd-41885.xml emd-41885.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41885_fsc.xml emd_41885_fsc.xml emd_41885_fsc_2.xml emd_41885_fsc_2.xml | 13.4 KB 17.7 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_41885.png emd_41885.png | 50 KB | ||
| Masks |  emd_41885_msk_1.map emd_41885_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41885.cif.gz emd-41885.cif.gz | 7.1 KB | ||
| Others |  emd_41885_half_map_1.map.gz emd_41885_half_map_1.map.gz emd_41885_half_map_2.map.gz emd_41885_half_map_2.map.gz | 200.6 MB 200.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41885 http://ftp.pdbj.org/pub/emdb/structures/EMD-41885 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41885 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41885 | HTTPS FTP |
-Validation report
| Summary document |  emd_41885_validation.pdf.gz emd_41885_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41885_full_validation.pdf.gz emd_41885_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_41885_validation.xml.gz emd_41885_validation.xml.gz | 21.8 KB | Display | |
| Data in CIF |  emd_41885_validation.cif.gz emd_41885_validation.cif.gz | 28.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41885 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41885 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41885 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41885 | HTTPS FTP |
-Related structure data
| Related structure data |  8u4kMC  8u4iC  8u4jC  8u4lC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41885.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41885.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
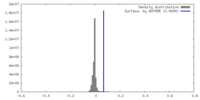
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41885_msk_1.map emd_41885_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
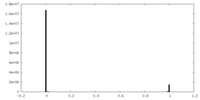
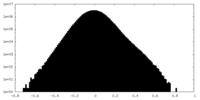
| Density Histograms |
-Half map: #2
| File | emd_41885_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
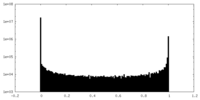
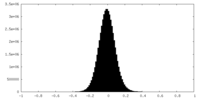
| Density Histograms |
-Half map: #1
| File | emd_41885_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
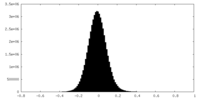
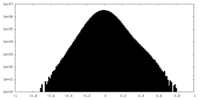
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of HER2/HER4/BTC
| Entire | Name: Ternary complex of HER2/HER4/BTC |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of HER2/HER4/BTC
| Supramolecule | Name: Ternary complex of HER2/HER4/BTC / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: HER2 and HER4 Receptors were expressed in EXPI293F cells. The ligand BTC was expressed in E. coli Origami B (DE3) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 284.435 KDa |
-Macromolecule #1: Isoform JM-A CYT-1 of Receptor tyrosine-protein kinase erbB-4
| Macromolecule | Name: Isoform JM-A CYT-1 of Receptor tyrosine-protein kinase erbB-4 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: receptor protein-tyrosine kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 67.909648 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVCAGTENK LSSLSDLEQQ YRALRKYYEN CEVVMGNLEI TSIEHNRDLS FLRSVREVTG YVLVALNQFR YLPLENLRII RGTKLYEDR YALAIFLNYR KDGNFGLQEL GLKNLTEILN GGVYVDQNKF LCYADTIHWQ DIVRNPWPSN LTLVSTNGSS G CGRCHKSC ...String: QSVCAGTENK LSSLSDLEQQ YRALRKYYEN CEVVMGNLEI TSIEHNRDLS FLRSVREVTG YVLVALNQFR YLPLENLRII RGTKLYEDR YALAIFLNYR KDGNFGLQEL GLKNLTEILN GGVYVDQNKF LCYADTIHWQ DIVRNPWPSN LTLVSTNGSS G CGRCHKSC TGRCWGPTEN HCQTLTRTVC AEQCDGRCYG PYVSDCCHRE CAGGCSGPKD TDCFACMNFN DSGACVTQCP QT FVYNPTT FQLEHNFNAK YTYGAFCVKK CPHNFVVDSS SCVRACPSSK MEVEENGIKM CKPCTDICPK ACDGIGTGSL MSA QTVDSS NIDKFINCTK INGNLIFLVT GIHGDPYNAI EAIDPEKLNV FRTVREITGF LNIQSWPPNM TDFSVFSNLV TIGG RVLYS GLSLLILKQQ GITSLQFQSL KEISAGNIYI TDNSNLCYYH TINWTTLFST INQRIVIRDN RKAENCTAEG MVCNH LCSS DGCWGPGPDQ CLSCRRFSRG RICIESCNLY DGEFREFENG SICVECDPQC EKMEDGLLTC HGPGPDNCTK CSHFKD GPN CVEKCPDGLQ GANSFIFKYA DPDRECHPCH PNCTQGCNGP TSHDCI UniProtKB: Receptor tyrosine-protein kinase erbB-4 |
-Macromolecule #2: Receptor tyrosine-protein kinase erbB-2
| Macromolecule | Name: Receptor tyrosine-protein kinase erbB-2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: receptor protein-tyrosine kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 66.888008 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVCTGTDMKL RLPASPETHL DMLRHLYQGC QVVQGNLELT YLPTNASLSF LQDIQEVQGY VLIAHNQVRQ VPLQRLRIVR GTQLFEDNY ALAVLDNGDP LNNTTPVTGA SPGGLRELQL RSLTEILKGG VLIQRNPQLC YQDTILWKDI FHKNNQLALT L IDTNRSRA ...String: QVCTGTDMKL RLPASPETHL DMLRHLYQGC QVVQGNLELT YLPTNASLSF LQDIQEVQGY VLIAHNQVRQ VPLQRLRIVR GTQLFEDNY ALAVLDNGDP LNNTTPVTGA SPGGLRELQL RSLTEILKGG VLIQRNPQLC YQDTILWKDI FHKNNQLALT L IDTNRSRA CHPCSPMCKG SRCWGESSED CQSLTRTVCA GGCARCKGPL PTDCCHEQCA AGCTGPKHSD CLACLHFNHS GI CELHCPA LVTYNTDTFE SMPNPEGRYT FGASCVTACP YNYLSTDVGS CTLVCPLHNQ EVTAEDGTQR CEKCSKPCAR VCY GLGMEH LREVRAVTSA NIQEFAGCKK IFGSLAFLPE SFDGDPASNT APLQPEQLQV FETLEEITGY LYISAWPDSL PDLS VFQNL QVIRGRILHN GAYSLTLQGL GISWLGLRSL RELGSGLALI HHNTHLCFVH TVPWDQLFRN PHQALLHTAN RPEDE CVGE GLACHQLCAR GHCWGPGPTQ CVNCSQFLRG QECVEECRVL QGLPREYVNA RHCLPCHPEC QPQNGSVTCF GPEADQ CVA CAHYKDPPFC VARCPSGVKP DLSYMPIWKF PDEEGACQPC PIN UniProtKB: Receptor tyrosine-protein kinase erbB-2 |
-Macromolecule #3: Betacellulin
| Macromolecule | Name: Betacellulin / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 5.630513 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GHFSRCPKQY KHYCIKGRCR FVVAEQTPSC VCDEGYIGAR CERVDLFY UniProtKB: Probetacellulin |
-Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 9 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: GRAPHENE OXIDE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average electron dose: 45.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)