[English] 日本語
 Yorodumi
Yorodumi- EMDB-41234: Complex of NPR1 ectodomain and REGN5381 Fab in an active-like sta... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
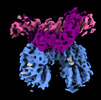
| Title | Complex of NPR1 ectodomain and REGN5381 Fab in an active-like state with no ANP bound | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Atrial natriuretic peptide / antibody / activator / pulmonary hypertension / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationANPR-A receptor complex / natriuretic peptide receptor activity / : / receptor guanylyl cyclase signaling pathway / body fluid secretion / peptide receptor activity / guanylate cyclase / cGMP biosynthetic process / guanylate cyclase activity / Physiological factors ...ANPR-A receptor complex / natriuretic peptide receptor activity / : / receptor guanylyl cyclase signaling pathway / body fluid secretion / peptide receptor activity / guanylate cyclase / cGMP biosynthetic process / guanylate cyclase activity / Physiological factors / positive regulation of renal sodium excretion / regulation of vascular permeability / G protein-coupled peptide receptor activity / positive regulation of urine volume / : / dopamine metabolic process / hormone binding / peptide hormone binding / negative regulation of angiogenesis / blood vessel diameter maintenance / negative regulation of smooth muscle cell proliferation / negative regulation of cell growth / regulation of blood pressure / cell surface receptor signaling pathway / protein kinase activity / receptor complex / GTP binding / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.65 Å | |||||||||
 Authors Authors | Franklin MC / Romero Hernandez A | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Agonist antibody to guanylate cyclase receptor NPR1 regulates vascular tone. Authors: Michael E Dunn / Aaron Kithcart / Jee Hae Kim / Andre Jo-Hao Ho / Matthew C Franklin / Annabel Romero Hernandez / Jan de Hoon / Wouter Botermans / Jonathan Meyer / Ximei Jin / Dongqin Zhang ...Authors: Michael E Dunn / Aaron Kithcart / Jee Hae Kim / Andre Jo-Hao Ho / Matthew C Franklin / Annabel Romero Hernandez / Jan de Hoon / Wouter Botermans / Jonathan Meyer / Ximei Jin / Dongqin Zhang / Justin Torello / Daniel Jasewicz / Vishal Kamat / Elena Garnova / Nina Liu / Michael Rosconi / Hao Pan / Satyajit Karnik / Michael E Burczynski / Wenjun Zheng / Ashique Rafique / Jonas B Nielsen / Tanima De / Niek Verweij / Anita Pandit / Adam Locke / Naga Chalasani / Olle Melander / Tae-Hwi Schwantes-An / / Aris Baras / Luca A Lotta / Bret J Musser / Jason Mastaitis / Kishor B Devalaraja-Narashimha / Andrew J Rankin / Tammy Huang / Gary Herman / William Olson / Andrew J Murphy / George D Yancopoulos / Benjamin A Olenchock / Lori Morton /    Abstract: Heart failure is a leading cause of morbidity and mortality. Elevated intracardiac pressures and myocyte stretch in heart failure trigger the release of counter-regulatory natriuretic peptides, which ...Heart failure is a leading cause of morbidity and mortality. Elevated intracardiac pressures and myocyte stretch in heart failure trigger the release of counter-regulatory natriuretic peptides, which act through their receptor (NPR1) to affect vasodilation, diuresis and natriuresis, lowering venous pressures and relieving venous congestion. Recombinant natriuretic peptide infusions were developed to treat heart failure but have been limited by a short duration of effect. Here we report that in a human genetic analysis of over 700,000 individuals, lifelong exposure to coding variants of the NPR1 gene is associated with changes in blood pressure and risk of heart failure. We describe the development of REGN5381, an investigational monoclonal agonist antibody that targets the membrane-bound guanylate cyclase receptor NPR1. REGN5381, an allosteric agonist of NPR1, induces an active-like receptor conformation that results in haemodynamic effects preferentially on venous vasculature, including reductions in systolic blood pressure and venous pressure in animal models. In healthy human volunteers, REGN5381 produced the expected haemodynamic effects, reflecting reductions in venous pressures, without obvious changes in diuresis and natriuresis. These data support the development of REGN5381 for long-lasting and selective lowering of venous pressures that drive symptomatology in patients with heart failure. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41234.map.gz emd_41234.map.gz | 107.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41234-v30.xml emd-41234-v30.xml emd-41234.xml emd-41234.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41234.png emd_41234.png | 123 KB | ||
| Filedesc metadata |  emd-41234.cif.gz emd-41234.cif.gz | 6.4 KB | ||
| Others |  emd_41234_half_map_1.map.gz emd_41234_half_map_1.map.gz emd_41234_half_map_2.map.gz emd_41234_half_map_2.map.gz | 105.6 MB 105.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41234 http://ftp.pdbj.org/pub/emdb/structures/EMD-41234 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41234 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41234 | HTTPS FTP |
-Related structure data
| Related structure data |  8tgaMC  8tg9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41234.map.gz / Format: CCP4 / Size: 113.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41234.map.gz / Format: CCP4 / Size: 113.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
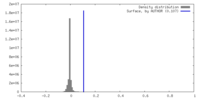
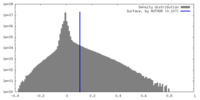
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_41234_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
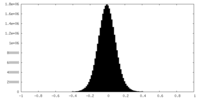
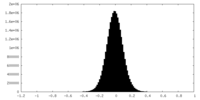
| Density Histograms |
-Half map: #2
| File | emd_41234_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
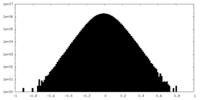
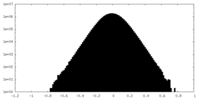
| Density Histograms |
- Sample components
Sample components
-Entire : Active-like complex of NPR1 ectodomain and REGN5381 Fab, with no ...
| Entire | Name: Active-like complex of NPR1 ectodomain and REGN5381 Fab, with no ANP present |
|---|---|
| Components |
|
-Supramolecule #1: Active-like complex of NPR1 ectodomain and REGN5381 Fab, with no ...
| Supramolecule | Name: Active-like complex of NPR1 ectodomain and REGN5381 Fab, with no ANP present type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Atrial natriuretic peptide receptor 1
| Macromolecule | Name: Atrial natriuretic peptide receptor 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: guanylate cyclase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.263723 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GNLTVAVVLP LANTSYPWSW ARVGPAVELA LAQVKARPDL LPGWTVRTVL GSSENALGVC SDTAAPLAAV DLKWEHNPAV FLGPGCVYA AAPVGRFTAH WRVPLLTAGA PALGFGVKDE YALTTRAGPS YAKLGDFVAA LHRRLGWERQ ALMLYAYRPG D EEHCFFLV ...String: GNLTVAVVLP LANTSYPWSW ARVGPAVELA LAQVKARPDL LPGWTVRTVL GSSENALGVC SDTAAPLAAV DLKWEHNPAV FLGPGCVYA AAPVGRFTAH WRVPLLTAGA PALGFGVKDE YALTTRAGPS YAKLGDFVAA LHRRLGWERQ ALMLYAYRPG D EEHCFFLV EGLFMRVRDR LNITVDHLEF AEDDLSHYTR LLRTMPRKGR VIYICSSPDA FRTLMLLALE AGLCGEDYVF FH LDIFGQS LQGGQGPAPR RPWERGDGQD VSARQAFQAA KIITYKDPDN PEYLEFLKQL KHLAYEQFNF TMEDGLVNTI PAS FHDGLL LYIQAVTETL AHGGTVTDGE NITQRMWNRS FQGVTGYLKI DSSGDRETDF SLWDMDPENG AFRVVLNYNG TSQE LVAVS GRKLNWPLGY PPPDIPKCGF DNEDPACNQD HLSTLEEQKL ISEEDLGGEQ KLISEEDLHH HHHH UniProtKB: Atrial natriuretic peptide receptor 1 |
-Macromolecule #2: REGN5381 Fab light chain
| Macromolecule | Name: REGN5381 Fab light chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.323898 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NIQMTQSPSS LSASVGDRVT ITCRASQSID SYLNWYQQKP GKAPKLLIYV ASSLQSGVPS RFSGSGSGKD FTLTISSLQP EDFATYYCQ QSYSIPTFGQ GTRLEIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ E SVTEQDSK ...String: NIQMTQSPSS LSASVGDRVT ITCRASQSID SYLNWYQQKP GKAPKLLIYV ASSLQSGVPS RFSGSGSGKD FTLTISSLQP EDFATYYCQ QSYSIPTFGQ GTRLEIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ E SVTEQDSK DSTYSLSSTL TLSKADYEKH KVYACEVTHQ GLSSPVTKSF NRGEC |
-Macromolecule #3: REGN5381 Fab heavy chain
| Macromolecule | Name: REGN5381 Fab heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.81877 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLVQSGAE VKKPGASVTV SCKASGYTFT DYYMHWVRQA PGQGLEWMGW IKPNSGGTNS AQRFQGRITM TWDTSISTAY MELSRLRSD DTAVYYCSRG GPVMNYYYYY GMDVWGQGTT VTVSSASTKG PSVFPLAPCS RSTSESTAAL GCLVKDYFPE P VTVSWNSG ...String: QVQLVQSGAE VKKPGASVTV SCKASGYTFT DYYMHWVRQA PGQGLEWMGW IKPNSGGTNS AQRFQGRITM TWDTSISTAY MELSRLRSD DTAVYYCSRG GPVMNYYYYY GMDVWGQGTT VTVSSASTKG PSVFPLAPCS RSTSESTAAL GCLVKDYFPE P VTVSWNSG ALTSGVHTFP AVLQSSGLYS LSSVVTVPSS SLGTKTYTCN VDHKPSNTKV DKRVESKYGP P |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.6 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 41.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)