+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4112 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Aichi virus 1: empty particle | |||||||||
 Map data Map data | Archie Virus 1: Empty particle | |||||||||
 Sample Sample | Aichi != Aichi virus 1 Aichi
| |||||||||
 Keywords Keywords | picornavirus / picornaviridae / Aichi virus 1 / empty particle / kobuvirus / 50 genome / release / human / pathogen / virus / RNA | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell Golgi membrane / symbiont-mediated suppression of host mRNA export from nucleus / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / channel activity / monoatomic ion transmembrane transport / RNA helicase activity / RNA helicase / RNA-directed RNA polymerase ...host cell Golgi membrane / symbiont-mediated suppression of host mRNA export from nucleus / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / channel activity / monoatomic ion transmembrane transport / RNA helicase activity / RNA helicase / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / symbiont entry into host cell / DNA-templated transcription / virion attachment to host cell / structural molecule activity / ATP hydrolysis activity / proteolysis / RNA binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  Aichi virus / Aichi virus /  Aichi virus 1 Aichi virus 1 | |||||||||
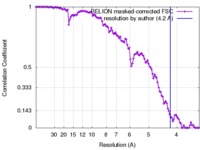
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Sabin C / Fuzik T | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2016 Journal: J Virol / Year: 2016Title: Structure of Aichi Virus 1 and Its Empty Particle: Clues to Kobuvirus Genome Release Mechanism. Authors: Charles Sabin / Tibor Füzik / Karel Škubník / Lenka Pálková / A Michael Lindberg / Pavel Plevka /   Abstract: (AiV-1) is a human pathogen from the genus of the family. Worldwide, 80 to 95% of adults have antibodies against the virus. AiV-1 infections are associated with nausea, gastroenteritis, and fever. ... (AiV-1) is a human pathogen from the genus of the family. Worldwide, 80 to 95% of adults have antibodies against the virus. AiV-1 infections are associated with nausea, gastroenteritis, and fever. Unlike most picornaviruses, kobuvirus capsids are composed of only three types of subunits: VP0, VP1, and VP3. We present here the structure of the AiV-1 virion determined to a resolution of 2.1 Å using X-ray crystallography. The surface loop puff of VP0 and knob of VP3 in AiV-1 are shorter than those in other picornaviruses. Instead, the 42-residue BC loop of VP0 forms the most prominent surface feature of the AiV-1 virion. We determined the structure of AiV-1 empty particle to a resolution of 4.2 Å using cryo-electron microscopy. The empty capsids are expanded relative to the native virus. The N-terminal arms of capsid proteins VP0, which mediate contacts between the pentamers of capsid protein protomers in the native AiV-1 virion, are disordered in the empty capsid. Nevertheless, the empty particles are stable, at least , and do not contain pores that might serve as channels for genome release. Therefore, extensive and probably reversible local reorganization of AiV-1 capsid is required for its genome release. Aichi virus 1 (AiV-1) is a human pathogen that can cause diarrhea, abdominal pain, nausea, vomiting, and fever. AiV-1 is identified in environmental screening studies with higher frequency and greater abundance than other human enteric viruses. Accordingly, 80 to 95% of adults worldwide have suffered from AiV-1 infections. We determined the structure of the AiV-1 virion. Based on the structure, we show that antiviral compounds that were developed against related enteroviruses are unlikely to be effective against AiV-1. The surface of the AiV-1 virion has a unique topology distinct from other related viruses from the family. We also determined that AiV-1 capsids form compact shells even after genome release. Therefore, AiV-1 genome release requires large localized and probably reversible reorganization of the capsid. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4112.map.gz emd_4112.map.gz | 8.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4112-v30.xml emd-4112-v30.xml emd-4112.xml emd-4112.xml | 11.8 KB 11.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4112_fsc.xml emd_4112_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_4112.png emd_4112.png | 170.7 KB | ||
| Filedesc metadata |  emd-4112.cif.gz emd-4112.cif.gz | 5.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4112 http://ftp.pdbj.org/pub/emdb/structures/EMD-4112 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4112 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4112 | HTTPS FTP |
-Related structure data
| Related structure data |  5lvcMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4112.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4112.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Archie Virus 1: Empty particle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.73 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Aichi
| Entire | Name: Aichi |
|---|---|
| Components |
|
-Supramolecule #1: Aichi virus 1
| Supramolecule | Name: Aichi virus 1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 1313215 / Sci species name: Aichi virus 1 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: VP1
| Macromolecule | Name: VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Aichi virus Aichi virus |
| Molecular weight | Theoretical: 27.194688 KDa |
| Recombinant expression | Organism:  Chlorocebus sabaeus (green monkey) Chlorocebus sabaeus (green monkey) |
| Sequence | String: TLTEDLDAPQ DTGNIENGAA DNSPQPRTTF DYTGNPLPPD TKLENFFSFY RLLPMGGSGA PSLSFPADEG TIIPLNPINW LKGADVSGI AAMLSCFTYI AADLRITLRF SNPNDNPATM LVAFAPPGAT IPLKPTRQML SNFYMAEVPV SAATSTMVSF S IPYTSPLS ...String: TLTEDLDAPQ DTGNIENGAA DNSPQPRTTF DYTGNPLPPD TKLENFFSFY RLLPMGGSGA PSLSFPADEG TIIPLNPINW LKGADVSGI AAMLSCFTYI AADLRITLRF SNPNDNPATM LVAFAPPGAT IPLKPTRQML SNFYMAEVPV SAATSTMVSF S IPYTSPLS AIPTSYFGWE DWSGTNFGQL SSGSWGNLML IPSLSVDSAI PFDFQLSCWV AFGNFKAWVP RPPPPLPPLP TP AANAERT VAVIKQ UniProtKB: Genome polyprotein |
-Macromolecule #2: VP0
| Macromolecule | Name: VP0 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Aichi virus Aichi virus |
| Molecular weight | Theoretical: 38.950758 KDa |
| Recombinant expression | Organism:  Chlorocebus sabaeus (green monkey) Chlorocebus sabaeus (green monkey) |
| Sequence | String: GNSVTNIYGN GNNVTTDVGA NGWAPTVSTG LGDGPVSASA DSLPGRSGGA SSEKTHTVSG SSNKVGSRFS KWWEPAAARA SESATDSAI EGIDAAGKAA SKAITRKLDR PAAPSSTANP QPSLIALNPS ATQSGNASIL TGSTAPSLLA YPTATPVPLP N PDEPSQPG ...String: GNSVTNIYGN GNNVTTDVGA NGWAPTVSTG LGDGPVSASA DSLPGRSGGA SSEKTHTVSG SSNKVGSRFS KWWEPAAARA SESATDSAI EGIDAAGKAA SKAITRKLDR PAAPSSTANP QPSLIALNPS ATQSGNASIL TGSTAPSLLA YPTATPVPLP N PDEPSQPG PSGDRTWLLD TVTWSQEFTR GWNIAGSNGM QWTGLESLIF PVSTDTNWTS TSSPTAYPLP FSFVRAYPDS SW AAMYNTH SMWNCGWRVQ VTVNGSQFHA GALILYMVPE ATTHAIQTAR DNAGFVFPYV ILNLYESNTA TIEVPYISPT PNT SSGLHA PWTFYLQVLS PLNPPPSLPT SLSCSIYVTP VDSSFHGLRY LAPQ UniProtKB: Genome polyprotein |
-Macromolecule #3: VP3
| Macromolecule | Name: VP3 / type: protein_or_peptide / ID: 3 / Details: VP3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Aichi virus Aichi virus |
| Molecular weight | Theoretical: 24.082244 KDa |
| Sequence | String: HWKTRAVPGA GTFGSAVAGQ ELPLCGVRAY YPPNAYIPAQ VRDWLEFAHR PGLMATVPWT MADEPAERLG IFPVSPSAIA GTGAPISYV ISLFSQWRGE LAAHLLFTGS AQHYGRLVVC YTPAAPQPPS TMQEAMRGTY TVWDVNAAST LEFTIPFISN S YWKTVDVN ...String: HWKTRAVPGA GTFGSAVAGQ ELPLCGVRAY YPPNAYIPAQ VRDWLEFAHR PGLMATVPWT MADEPAERLG IFPVSPSAIA GTGAPISYV ISLFSQWRGE LAAHLLFTGS AQHYGRLVVC YTPAAPQPPS TMQEAMRGTY TVWDVNAAST LEFTIPFISN S YWKTVDVN NPDALLSTTG YVSIWVQNPL VGPHTAPASA LVQAFISAGE SFNVRLMQNP ALTSQ UniProtKB: Genome polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)