+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of dodecameric hub domain of CaMKII alpha | |||||||||
 Map data Map data | Dodecameric CaMKII alpha hub | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | High-order oligomer / Protein Kinase / Signaling / Memory / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of synaptic vesicle docking / HSF1-dependent transactivation / RAF activation / Ion transport by P-type ATPases / peptidyl-threonine autophosphorylation / calcium- and calmodulin-dependent protein kinase complex / neurotransmitter receptor transport to plasma membrane / regulation of endocannabinoid signaling pathway / Interferon gamma signaling / Ca2+/calmodulin-dependent protein kinase ...regulation of synaptic vesicle docking / HSF1-dependent transactivation / RAF activation / Ion transport by P-type ATPases / peptidyl-threonine autophosphorylation / calcium- and calmodulin-dependent protein kinase complex / neurotransmitter receptor transport to plasma membrane / regulation of endocannabinoid signaling pathway / Interferon gamma signaling / Ca2+/calmodulin-dependent protein kinase / dendritic spine development / negative regulation of hydrolase activity / regulation of neurotransmitter secretion / Trafficking of AMPA receptors / regulation of neuron migration / positive regulation of calcium ion transport / calcium/calmodulin-dependent protein kinase activity / Ca2+ pathway / GTPase activating protein binding / regulation of mitochondrial membrane permeability involved in apoptotic process / RAF/MAP kinase cascade / Ion homeostasis / dendrite morphogenesis / regulation of neurotransmitter receptor localization to postsynaptic specialization membrane / regulation of neuronal synaptic plasticity / Unblocking of NMDA receptors, glutamate binding and activation / glutamate receptor binding / postsynaptic cytosol / positive regulation of cardiac muscle cell apoptotic process / regulation of protein localization to plasma membrane / presynaptic cytosol / cellular response to interferon-beta / ionotropic glutamate receptor signaling pathway / dendrite cytoplasm / bioluminescence / response to ischemia / angiotensin-activated signaling pathway / positive regulation of receptor signaling pathway via JAK-STAT / generation of precursor metabolites and energy / G1/S transition of mitotic cell cycle / cellular response to type II interferon / Schaffer collateral - CA1 synapse / kinase activity / calcium ion transport / dendritic spine / calmodulin binding / neuron projection / postsynaptic density / axon / protein serine kinase activity / protein serine/threonine kinase activity / neuronal cell body / dendrite / synapse / glutamatergic synapse / protein homodimerization activity / mitochondrion / ATP binding / metal ion binding / identical protein binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Chien C-T / Chiu W / Khan S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2024 Journal: Commun Biol / Year: 2024Title: Hub stability in the calcium calmodulin-dependent protein kinase II. Authors: Chih-Ta Chien / Henry Puhl / Steven S Vogel / Justin E Molloy / Wah Chiu / Shahid Khan /   Abstract: The calcium calmodulin protein kinase II (CaMKII) is a multi-subunit ring assembly with a central hub formed by the association domains. There is evidence for hub polymorphism between and within ...The calcium calmodulin protein kinase II (CaMKII) is a multi-subunit ring assembly with a central hub formed by the association domains. There is evidence for hub polymorphism between and within CaMKII isoforms, but the link between polymorphism and subunit exchange has not been resolved. Here, we present near-atomic resolution cryogenic electron microscopy (cryo-EM) structures revealing that hubs from the α and β isoforms, either standalone or within an β holoenzyme, coexist as 12 and 14 subunit assemblies. Single-molecule fluorescence microscopy of Venus-tagged holoenzymes detects intermediate assemblies and progressive dimer loss due to intrinsic holoenzyme lability, and holoenzyme disassembly into dimers upon mutagenesis of a conserved inter-domain contact. Molecular dynamics (MD) simulations show the flexibility of 4-subunit precursors, extracted in-silico from the β hub polymorphs, encompassing the curvature of both polymorphs. The MD explains how an open hub structure also obtained from the β holoenzyme sample could be created by dimer loss and analysis of its cryo-EM dataset reveals how the gap could open further. An assembly model, considering dimer concentration dependence and strain differences between polymorphs, proposes a mechanism for intrinsic hub lability to fine-tune the stoichiometry of αβ heterooligomers for their dynamic localization within synapses in neurons. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40955.map.gz emd_40955.map.gz | 203.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40955-v30.xml emd-40955-v30.xml emd-40955.xml emd-40955.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40955.png emd_40955.png | 62.6 KB | ||
| Filedesc metadata |  emd-40955.cif.gz emd-40955.cif.gz | 5.5 KB | ||
| Others |  emd_40955_half_map_1.map.gz emd_40955_half_map_1.map.gz emd_40955_half_map_2.map.gz emd_40955_half_map_2.map.gz | 200.1 MB 200.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40955 http://ftp.pdbj.org/pub/emdb/structures/EMD-40955 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40955 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40955 | HTTPS FTP |
-Related structure data
| Related structure data |  8t15MC  8sygC  8t17C  8t18C  8t6kC  8t6qC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40955.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40955.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Dodecameric CaMKII alpha hub | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.741 Å | ||||||||||||||||||||||||||||||||||||
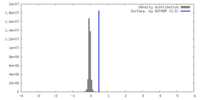
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_40955_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
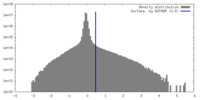
| Density Histograms |
-Half map: #1
| File | emd_40955_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Venus-tagged CaMKII Alpha Association Domain
| Entire | Name: Venus-tagged CaMKII Alpha Association Domain |
|---|---|
| Components |
|
-Supramolecule #1: Venus-tagged CaMKII Alpha Association Domain
| Supramolecule | Name: Venus-tagged CaMKII Alpha Association Domain / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Venus-tagged CaMKII Alpha Association Domain
| Macromolecule | Name: Venus-tagged CaMKII Alpha Association Domain / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO / EC number: Ca2+/calmodulin-dependent protein kinase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.877539 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH VSKGEELFTG VVPILVELDG DVNGHKFSVS GEGEGDATYG KLTLKLICTT GKLPVPWPTL VTTLGYGLQ CFARYPDHMK QHDFFKSAMP EGYVQERTIF FKDDGNYKTR AEVKFEGDTL VNRIELKGID FKEDGNILGH K LEYNYNSH ...String: MGSSHHHHHH SSGLVPRGSH VSKGEELFTG VVPILVELDG DVNGHKFSVS GEGEGDATYG KLTLKLICTT GKLPVPWPTL VTTLGYGLQ CFARYPDHMK QHDFFKSAMP EGYVQERTIF FKDDGNYKTR AEVKFEGDTL VNRIELKGID FKEDGNILGH K LEYNYNSH NVYITADKQK NGIKANFKIR HNIEDGGVQL ADHYQQNTPI GDGPVLLPDN HYLSYQSKLS KDPNEKRDHM VL LEFVTAA GITLGMDELY KSGLRSRAQA SNSAVDVRKQ EIIKVTEQLI EAISNGDFES YTKMCDPGMT AFEPEALGNL VEG LDFHRF YFENLWSRNS KPVHTTILNP HIHLMGDESA CIAYIRITQY LDAGGIPRTA QSEETRVWHR RDGKWQIVHF HRSG APSVL PH UniProtKB: Green fluorescent protein, Calcium/calmodulin-dependent protein kinase type II subunit alpha |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 20 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 94506 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)