[English] 日本語
 Yorodumi
Yorodumi- EMDB-3892: State F (Rix1-TAP Rpf2-Flag) - Visualizing the assembly pathway o... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3892 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | State F (Rix1-TAP Rpf2-Flag) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | |||||||||
 Map data Map data | State F (Rix1-TAP Rpf2-Flag) - Visualizing the assembly pathway of nucleolar pre-60S ribosmes | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
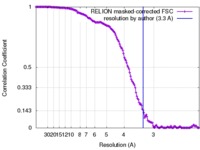
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Kater L / Thoms M / Barrio-Garcia C / Cheng J / Ismail S / Ahmed YL / Bange G / Kressler D / Berninghausen O / Sinning I ...Kater L / Thoms M / Barrio-Garcia C / Cheng J / Ismail S / Ahmed YL / Bange G / Kressler D / Berninghausen O / Sinning I / Hurt E / Beckmann R | |||||||||
 Citation Citation |  Journal: Cell / Year: 2017 Journal: Cell / Year: 2017Title: Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes. Authors: Lukas Kater / Matthias Thoms / Clara Barrio-Garcia / Jingdong Cheng / Sherif Ismail / Yasar Luqman Ahmed / Gert Bange / Dieter Kressler / Otto Berninghausen / Irmgard Sinning / Ed Hurt / Roland Beckmann /  Abstract: Eukaryotic 60S ribosomal subunits are comprised of three rRNAs and ∼50 ribosomal proteins. The initial steps of their formation take place in the nucleolus, but, owing to a lack of structural ...Eukaryotic 60S ribosomal subunits are comprised of three rRNAs and ∼50 ribosomal proteins. The initial steps of their formation take place in the nucleolus, but, owing to a lack of structural information, this process is poorly understood. Using cryo-EM, we solved structures of early 60S biogenesis intermediates at 3.3 Å to 4.5 Å resolution, thereby providing insights into their sequential folding and assembly pathway. Besides revealing distinct immature rRNA conformations, we map 25 assembly factors in six different assembly states. Notably, the Nsa1-Rrp1-Rpf1-Mak16 module stabilizes the solvent side of the 60S subunit, and the Erb1-Ytm1-Nop7 complex organizes and connects through Erb1's meandering N-terminal extension, eight assembly factors, three ribosomal proteins, and three 25S rRNA domains. Our structural snapshots reveal the order of integration and compaction of the six major 60S domains within early nucleolar 60S particles developing stepwise from the solvent side around the exit tunnel to the central protuberance. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3892.map.gz emd_3892.map.gz | 262.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3892-v30.xml emd-3892-v30.xml emd-3892.xml emd-3892.xml | 9.3 KB 9.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3892_fsc.xml emd_3892_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_3892.png emd_3892.png | 69.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3892 http://ftp.pdbj.org/pub/emdb/structures/EMD-3892 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3892 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3892 | HTTPS FTP |
-Validation report
| Summary document |  emd_3892_validation.pdf.gz emd_3892_validation.pdf.gz | 307.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3892_full_validation.pdf.gz emd_3892_full_validation.pdf.gz | 306.3 KB | Display | |
| Data in XML |  emd_3892_validation.xml.gz emd_3892_validation.xml.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3892 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3892 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3892 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3892 | HTTPS FTP |
-Related structure data
| Related structure data |  3888C  3889C  3890C  3891C  3893C  6elzC  6em1C  6em3C  6em4C  6em5C  6emfC  6emgC  6en7C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3892.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3892.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | State F (Rix1-TAP Rpf2-Flag) - Visualizing the assembly pathway of nucleolar pre-60S ribosmes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.084 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
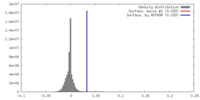
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Large subunit biogenesis particle purified via Rix1-TAP and Rpf2-Flag
| Entire | Name: Large subunit biogenesis particle purified via Rix1-TAP and Rpf2-Flag |
|---|---|
| Components |
|
-Supramolecule #1: Large subunit biogenesis particle purified via Rix1-TAP and Rpf2-Flag
| Supramolecule | Name: Large subunit biogenesis particle purified via Rix1-TAP and Rpf2-Flag type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 27.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)