[English] 日本語
 Yorodumi
Yorodumi- EMDB-38586: Structure 2 of human class T GPCR TAS2R14-miniGs/gust complex wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure 2 of human class T GPCR TAS2R14-miniGs/gust complex with Flufenamic acid. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR Tas2R14 miniGsg FLF Bitter receptor / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationbitter taste receptor activity / taste receptor activity / detection of chemical stimulus involved in sensory perception of bitter taste / Class C/3 (Metabotropic glutamate/pheromone receptors) / G protein-coupled receptor activity / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor ...bitter taste receptor activity / taste receptor activity / detection of chemical stimulus involved in sensory perception of bitter taste / Class C/3 (Metabotropic glutamate/pheromone receptors) / G protein-coupled receptor activity / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / protein-containing complex binding / signal transduction / extracellular exosome / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
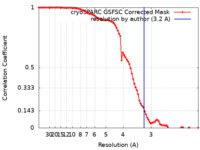
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Hu XL / Wu LJ / Hua T / Liu ZJ | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Bitter taste TAS2R14 activation by intracellular tastants and cholesterol. Authors: Xiaolong Hu / Weizhen Ao / Mingxin Gao / Lijie Wu / Yuan Pei / Shenhui Liu / Yiran Wu / Fei Zhao / Qianqian Sun / Junlin Liu / Longquan Jiang / Xin Wang / Yan Li / Qiwen Tan / Jie Cheng / ...Authors: Xiaolong Hu / Weizhen Ao / Mingxin Gao / Lijie Wu / Yuan Pei / Shenhui Liu / Yiran Wu / Fei Zhao / Qianqian Sun / Junlin Liu / Longquan Jiang / Xin Wang / Yan Li / Qiwen Tan / Jie Cheng / Fan Yang / Chi Yang / Jinpeng Sun / Tian Hua / Zhi-Jie Liu /  Abstract: Bitter taste receptors, particularly TAS2R14, play central roles in discerning a wide array of bitter substances, ranging from dietary components to pharmaceutical agents. TAS2R14 is also widely ...Bitter taste receptors, particularly TAS2R14, play central roles in discerning a wide array of bitter substances, ranging from dietary components to pharmaceutical agents. TAS2R14 is also widely expressed in extragustatory tissues, suggesting its extra roles in diverse physiological processes and potential therapeutic applications. Here we present cryogenic electron microscopy structures of TAS2R14 in complex with aristolochic acid, flufenamic acid and compound 28.1, coupling with different G-protein subtypes. Uniquely, a cholesterol molecule is observed occupying what is typically an orthosteric site in class A G-protein-coupled receptors. The three potent agonists bind, individually, to the intracellular pockets, suggesting a distinct activation mechanism for this receptor. Comprehensive structural analysis, combined with mutagenesis and molecular dynamic simulation studies, elucidate the broad-spectrum ligand recognition and activation of the receptor by means of intricate multiple ligand-binding sites. Our study also uncovers the specific coupling modes of TAS2R14 with gustducin and G proteins. These findings should be instrumental in advancing knowledge of bitter taste perception and its broader implications in sensory biology and drug discovery. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38586.map.gz emd_38586.map.gz | 56.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38586-v30.xml emd-38586-v30.xml emd-38586.xml emd-38586.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38586_fsc.xml emd_38586_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_38586.png emd_38586.png | 51.4 KB | ||
| Filedesc metadata |  emd-38586.cif.gz emd-38586.cif.gz | 7.5 KB | ||
| Others |  emd_38586_half_map_1.map.gz emd_38586_half_map_1.map.gz emd_38586_half_map_2.map.gz emd_38586_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38586 http://ftp.pdbj.org/pub/emdb/structures/EMD-38586 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38586 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38586 | HTTPS FTP |
-Related structure data
| Related structure data |  8xqrMC  8xqlC  8xqnC  8xqoC  8xqpC  8xqsC  8xqtC  8ykyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38586.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38586.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
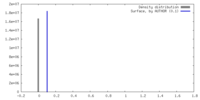
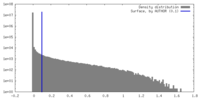
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_38586_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
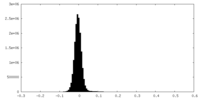
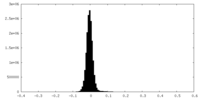
| Density Histograms |
-Half map: #1
| File | emd_38586_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
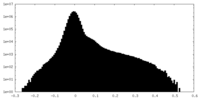
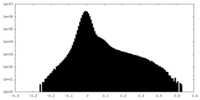
| Density Histograms |
- Sample components
Sample components
-Entire : Structure 2 of human class T GPCR TAS2R14-miniGs/gust complex wit...
| Entire | Name: Structure 2 of human class T GPCR TAS2R14-miniGs/gust complex with Flufenamic acid. |
|---|---|
| Components |
|
-Supramolecule #1: Structure 2 of human class T GPCR TAS2R14-miniGs/gust complex wit...
| Supramolecule | Name: Structure 2 of human class T GPCR TAS2R14-miniGs/gust complex with Flufenamic acid. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(t) subunit alpha-3
| Macromolecule | Name: Guanine nucleotide-binding protein G(t) subunit alpha-3 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 30.553307 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDYKDDDDKE NLYFQGNSKT EDQRNEEKAQ REANKKIEKQ LQKDKQVYRA THRLLLLGAD NSGKSTIVKQ MRILHGGSGG SGGTSGIFE TKFQVDKVNF HMFDVGGQRD ERRKWIQCFN DVTAIIFVVD SSDYNRLQEA LNDFKSIWNN RWLRTISVIL F LNKQDLLA ...String: MDYKDDDDKE NLYFQGNSKT EDQRNEEKAQ REANKKIEKQ LQKDKQVYRA THRLLLLGAD NSGKSTIVKQ MRILHGGSGG SGGTSGIFE TKFQVDKVNF HMFDVGGQRD ERRKWIQCFN DVTAIIFVVD SSDYNRLQEA LNDFKSIWNN RWLRTISVIL F LNKQDLLA EKVLAGKSKI EDYFPEFARY TTPEDATPEP GEDPRVTRAK YFIRDEFLRI STASGDGRHY CYPHFTCAVD TQ NVKFVFD AVTDIIIKEN LKDCGLF |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.728426 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWNGSSGGG GSGGGGSSGV SGWRLFKKIS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nanobody 35
| Macromolecule | Name: Nanobody 35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.845516 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQVQLQESGG GLVQPGGSLR LSCAASGFTF SNYKMNWVRQ APGKGLEWVS DISQSGASIS YTGSVKGRFT ISRDNAKNTL YLQMNSLKP EDTAVYYCAR CPAPFTRDCF DVTSTTYAYR GQGTQVTVSS HHHHHH |
-Macromolecule #5: Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT
| Macromolecule | Name: Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 110.840023 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAHHHHHH HHHHENLYFQ SGRAVEGAVK TEPVDLFHPG FLNSSNYRIP ALFKTKEGTL IASIDARRH GGADAPNNDI DTAVRRSEDG GKTWDEGQII MDYPDKSSVI DTTLIQDDET GRIFLLVTHF PSKYGFWNAG L GSGFKNID ...String: MKTIIALSYI FCLVFADYKD DDDAHHHHHH HHHHENLYFQ SGRAVEGAVK TEPVDLFHPG FLNSSNYRIP ALFKTKEGTL IASIDARRH GGADAPNNDI DTAVRRSEDG GKTWDEGQII MDYPDKSSVI DTTLIQDDET GRIFLLVTHF PSKYGFWNAG L GSGFKNID GKEYLCLYDS SGKEFTVREN VVYDKDSNKT EYTTNALGDL FKNGTKIDNI NSSTAPLKAK GTSYINLVYS DD DGKTWSE PQNINFQVKK DWMKFLGIAP GRGIQIKNGE HKGRIVVPVY YTNEKGKQSS AVIYSDDSGK NWTIGESPND NRK LENGKI INSKTLSDDA PQLTECQVVE MPNGQLKLFM RNLSGYLNIA TSFDGGATWD ETVEKDTNVL EPYCQLSVIN YSQK VDGKD AVIFSNPNAR SRSNGTVRIG LINQVGTYEN GEPKYEFDWK YNKLVKPGYY AYSCLTELSN GNIGLLYEGT PSEEM SYIE MNLKYLESGA NKGSAGSGGV IKSIFTFVLI VEFIIGNLGN SFIALVNCID WVKGRKISSV DRILTALAIS RISLVW LIF GSWCVSVFFP ALFATEKMFR MLTNIWTVIN HFSVWLATGL GTFYFLKIAN FSNSIFLYLK WRVKKVVLVL LLVTSVF LF LNIALINIHI NASINGYRRN KTCSSDSSNF TRFSSLIVLT STVFIFIPFT LSLAMFLLLI FSMWKHRKKM QHTVKISG D ASTKAHRGVK SVITFFLLYA IFSLSFFISV WTSERLEENL IILSQVMGMA YPSCHSCVLI LGNKKLRQAS LSVLLWLRY MFKDGEPSGH KEFRESSGSG SSGSGSSGSG SSVFTLEDFV GDWEQTAAYN LDQVLEQGGV SSLLQNLAVS VTPIQRIVRS GENALKIDI HVIIPYEGLS ADQMAQIEEV FKVVYPVDDH HFKVILPYGT LVIDGVTPNM LNYFGRPYEG IAVFDGKKIT V TGTLWNGN KIIDERLITP DGSMLFRVTI NS UniProtKB: UNIPROTKB: A0A6M1VLG5, Taste receptor type 2 member 14 |
-Macromolecule #6: 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID
| Macromolecule | Name: 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID / type: ligand / ID: 6 / Number of copies: 1 / Formula: FLF |
|---|---|
| Molecular weight | Theoretical: 281.23 Da |
| Chemical component information |  ChemComp-FLF: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)