+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | ||||||||||||||||||
 マップデータ マップデータ | |||||||||||||||||||
 試料 試料 |
| ||||||||||||||||||
 キーワード キーワード | Nucleosome / Recombinase / DNA BINDING PROTEIN-DNA Complex | ||||||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報presynaptic intermediate filament cytoskeleton / response to glucoside / mitotic recombination-dependent replication fork processing / chromosome organization involved in meiotic cell cycle / cellular response to camptothecin / DNA recombinase assembly / telomere maintenance via telomere lengthening / double-strand break repair involved in meiotic recombination / nuclear ubiquitin ligase complex / cellular response to cisplatin ...presynaptic intermediate filament cytoskeleton / response to glucoside / mitotic recombination-dependent replication fork processing / chromosome organization involved in meiotic cell cycle / cellular response to camptothecin / DNA recombinase assembly / telomere maintenance via telomere lengthening / double-strand break repair involved in meiotic recombination / nuclear ubiquitin ligase complex / cellular response to cisplatin / DNA strand invasion / cellular response to hydroxyurea / mitotic recombination / lateral element / DNA strand exchange activity / replication-born double-strand break repair via sister chromatid exchange / telomere maintenance via recombination / regulation of DNA damage checkpoint / Impaired BRCA2 binding to PALB2 / single-stranded DNA helicase activity / reciprocal meiotic recombination / Homologous DNA Pairing and Strand Exchange / Defective homologous recombination repair (HRR) due to BRCA1 loss of function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA1 binding function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA2/RAD51/RAD51C binding function / Resolution of D-loop Structures through Synthesis-Dependent Strand Annealing (SDSA) / Resolution of D-loop Structures through Holliday Junction Intermediates / HDR through Single Strand Annealing (SSA) / ATP-dependent DNA damage sensor activity / regulation of double-strand break repair via homologous recombination / nuclear chromosome / Impaired BRCA2 binding to RAD51 / Transcriptional Regulation by E2F6 / replication fork processing / Presynaptic phase of homologous DNA pairing and strand exchange / negative regulation of tumor necrosis factor-mediated signaling pathway / ATP-dependent activity, acting on DNA / response to X-ray / negative regulation of megakaryocyte differentiation / interstrand cross-link repair / protein localization to CENP-A containing chromatin / Chromatin modifying enzymes / Replacement of protamines by nucleosomes in the male pronucleus / CENP-A containing nucleosome / Packaging Of Telomere Ends / condensed chromosome / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / DNA polymerase binding / Recognition and association of DNA glycosylase with site containing an affected pyrimidine / Cleavage of the damaged pyrimidine / Deposition of new CENPA-containing nucleosomes at the centromere / telomere organization / Inhibition of DNA recombination at telomere / Meiotic synapsis / Interleukin-7 signaling / RNA Polymerase I Promoter Opening / Assembly of the ORC complex at the origin of replication / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / innate immune response in mucosa / SUMOylation of chromatin organization proteins / male germ cell nucleus / DNA methylation / Condensation of Prophase Chromosomes / Chromatin modifications during the maternal to zygotic transition (MZT) / HCMV Late Events / SIRT1 negatively regulates rRNA expression / epigenetic regulation of gene expression / condensed nuclear chromosome / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / PRC2 methylates histones and DNA / Regulation of endogenous retroelements by KRAB-ZFP proteins / Defective pyroptosis / meiotic cell cycle / cellular response to ionizing radiation / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / HDACs deacetylate histones / Nonhomologous End-Joining (NHEJ) / RNA Polymerase I Promoter Escape / lipopolysaccharide binding / Transcriptional regulation by small RNAs / Formation of the beta-catenin:TCF transactivating complex / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / double-strand break repair via homologous recombination / HDMs demethylate histones / G2/M DNA damage checkpoint / cellular response to gamma radiation / NoRC negatively regulates rRNA expression / PML body / DNA Damage/Telomere Stress Induced Senescence / B-WICH complex positively regulates rRNA expression / PKMTs methylate histone lysines / Meiotic recombination / HDR through Homologous Recombination (HRR) / Pre-NOTCH Transcription and Translation / Metalloprotease DUBs / RMTs methylate histone arginines / Activation of anterior HOX genes in hindbrain development during early embryogenesis / Transcriptional regulation of granulopoiesis 類似検索 - 分子機能 | ||||||||||||||||||
| 生物種 |  Homo sapiens (ヒト) / synthetic construct (人工物) Homo sapiens (ヒト) / synthetic construct (人工物) | ||||||||||||||||||
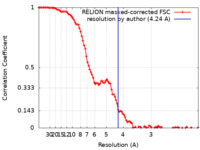
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 4.24 Å | ||||||||||||||||||
 データ登録者 データ登録者 | Shioi T / Hatazawa S / Ogasawara M / Takizawa Y / Kurumizaka H | ||||||||||||||||||
| 資金援助 |  日本, 5件 日本, 5件
| ||||||||||||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2024 ジャーナル: Nature / 年: 2024タイトル: Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site. 著者: Takuro Shioi / Suguru Hatazawa / Eriko Oya / Noriko Hosoya / Wataru Kobayashi / Mitsuo Ogasawara / Takehiko Kobayashi / Yoshimasa Takizawa / Hitoshi Kurumizaka /  要旨: RAD51 is the central eukaryotic recombinase required for meiotic recombination and mitotic repair of double-strand DNA breaks (DSBs). However, the mechanism by which RAD51 functions at DSB sites in ...RAD51 is the central eukaryotic recombinase required for meiotic recombination and mitotic repair of double-strand DNA breaks (DSBs). However, the mechanism by which RAD51 functions at DSB sites in chromatin has remained elusive. Here we report the cryo-electron microscopy structures of human RAD51-nucleosome complexes, in which RAD51 forms ring and filament conformations. In the ring forms, the N-terminal lobe domains (NLDs) of RAD51 protomers are aligned on the outside of the RAD51 ring, and directly bind to the nucleosomal DNA. The nucleosomal linker DNA that contains the DSB site is recognized by the L1 and L2 loops-active centres that face the central hole of the RAD51 ring. In the filament form, the nucleosomal DNA is peeled by the RAD51 filament extension, and the NLDs of RAD51 protomers proximal to the nucleosome bind to the remaining nucleosomal DNA and histones. Mutations that affect nucleosome-binding residues of the RAD51 NLD decrease nucleosome binding, but barely affect DNA binding in vitro. Consistently, yeast Rad51 mutants with the corresponding mutations are substantially defective in DNA repair in vivo. These results reveal an unexpected function of the RAD51 NLD, and explain the mechanism by which RAD51 associates with nucleosomes, recognizes DSBs and forms the active filament in chromatin. | ||||||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_38229.map.gz emd_38229.map.gz | 10.4 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-38229-v30.xml emd-38229-v30.xml emd-38229.xml emd-38229.xml | 21.7 KB 21.7 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_38229_fsc.xml emd_38229_fsc.xml | 10 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_38229.png emd_38229.png | 143.4 KB | ||
| Filedesc metadata |  emd-38229.cif.gz emd-38229.cif.gz | 6.6 KB | ||
| その他 |  emd_38229_half_map_1.map.gz emd_38229_half_map_1.map.gz emd_38229_half_map_2.map.gz emd_38229_half_map_2.map.gz | 65.5 MB 65.6 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38229 http://ftp.pdbj.org/pub/emdb/structures/EMD-38229 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38229 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38229 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_38229_validation.pdf.gz emd_38229_validation.pdf.gz | 822.9 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_38229_full_validation.pdf.gz emd_38229_full_validation.pdf.gz | 822.5 KB | 表示 | |
| XML形式データ |  emd_38229_validation.xml.gz emd_38229_validation.xml.gz | 17.1 KB | 表示 | |
| CIF形式データ |  emd_38229_validation.cif.gz emd_38229_validation.cif.gz | 22.5 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38229 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38229 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38229 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38229 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8xbuMC  8jndC  8jneC  8jnfC  8xbtC  8xbvC  8xbwC  8xbxC  8xbyC  36446 M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_38229.map.gz / 形式: CCP4 / 大きさ: 83.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_38229.map.gz / 形式: CCP4 / 大きさ: 83.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: #2
| ファイル | emd_38229_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
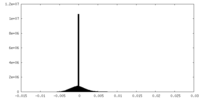
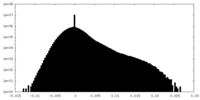
| 密度ヒストグラム |
-ハーフマップ: #1
| ファイル | emd_38229_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
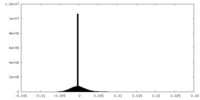
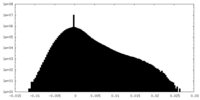
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : RAD51-nucleosome complex
| 全体 | 名称: RAD51-nucleosome complex |
|---|---|
| 要素 |
|
-超分子 #1: RAD51-nucleosome complex
| 超分子 | 名称: RAD51-nucleosome complex / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#4 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Histone H3.1
| 分子 | 名称: Histone H3.1 / タイプ: protein_or_peptide / ID: 1 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 15.719445 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: GSHMARTKQT ARKSTGGKAP RKQLATKAAR KSAPATGGVK KPHRYRPGTV ALREIRRYQK STELLIRKLP FQRLVREIAQ DFKTDLRFQ SSAVMALQEA CEAYLVGLFE DTNLCAIHAK RVTIMPKDIQ LARRIRGERA UniProtKB: Histone H3.1 |
-分子 #2: Histone H4
| 分子 | 名称: Histone H4 / タイプ: protein_or_peptide / ID: 2 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 11.676703 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: GSHMSGRGKG GKGLGKGGAK RHRKVLRDNI QGITKPAIRR LARRGGVKRI SGLIYEETRG VLKVFLENVI RDAVTYTEHA KRKTVTAMD VVYALKRQGR TLYGFGG UniProtKB: Histone H4 |
-分子 #3: Histone H2A type 1-B/E
| 分子 | 名称: Histone H2A type 1-B/E / タイプ: protein_or_peptide / ID: 3 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 14.447825 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: GSHMSGRGKQ GGKARAKAKT RSSRAGLQFP VGRVHRLLRK GNYSERVGAG APVYLAAVLE YLTAEILELA GNAARDNKKT RIIPRHLQL AIRNDEELNK LLGRVTIAQG GVLPNIQAVL LPKKTESHHK AKGK UniProtKB: Histone H2A type 1-B/E |
-分子 #4: Histone H2B type 1-J
| 分子 | 名称: Histone H2B type 1-J / タイプ: protein_or_peptide / ID: 4 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 14.217516 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: GSHMPEPAKS APAPKKGSKK AVTKAQKKDG KKRKRSRKES YSIYVYKVLK QVHPDTGISS KAMGIMNSFV NDIFERIAGE ASRLAHYNK RSTITSREIQ TAVRLLLPGE LAKHAVSEGT KAVTKYTSAK UniProtKB: Histone H2B type 1-J |
-分子 #7: DNA repair protein RAD51 homolog 1
| 分子 | 名称: DNA repair protein RAD51 homolog 1 / タイプ: protein_or_peptide / ID: 7 / コピー数: 10 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 37.291398 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: GSHMAMQMQL EANADTSVEE ESFGPQPISR LEQCGINAND VKKLEEAGFH TVEAVAYAPK KELINIKGIS EAKADKILAE AAKLVPMGF TTATEFHQRR SEIIQITTGS KELDKLLQGG IETGSITEMF GEFRTGKTQI CHTLAVTCQL PIDRGGGEGK A MYIDTEGT ...文字列: GSHMAMQMQL EANADTSVEE ESFGPQPISR LEQCGINAND VKKLEEAGFH TVEAVAYAPK KELINIKGIS EAKADKILAE AAKLVPMGF TTATEFHQRR SEIIQITTGS KELDKLLQGG IETGSITEMF GEFRTGKTQI CHTLAVTCQL PIDRGGGEGK A MYIDTEGT FRPERLLAVA ERYGLSGSDV LDNVAYARAF NTDHQTQLLY QASAMMVESR YALLIVDSAT ALYRTDYSGR GE LSARQMH LARFLRMLLR LADEFGVAVV ITNQVVAQVD GAAMFAADPK KPIGGNIIAH ASTTRLYLRK GRGETRICKI YDS PCLPEA EAMFAINADG VGDAKD UniProtKB: DNA repair protein RAD51 homolog 1 |
-分子 #5: DNA (156-MER)
| 分子 | 名称: DNA (156-MER) / タイプ: dna / ID: 5 / コピー数: 1 / 分類: DNA |
|---|---|
| 由来(天然) | 生物種: synthetic construct (人工物) |
| 分子量 | 理論値: 47.976699 KDa |
| 配列 | 文字列: (DA)(DT)(DC)(DA)(DG)(DA)(DA)(DT)(DC)(DC) (DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA)(DG) (DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA) (DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA)(DG) (DA) (DC)(DA)(DG)(DC)(DT) ...文字列: (DA)(DT)(DC)(DA)(DG)(DA)(DA)(DT)(DC)(DC) (DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA)(DG) (DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA) (DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA)(DG) (DA) (DC)(DA)(DG)(DC)(DT)(DC)(DT)(DA) (DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT)(DT)(DA) (DA)(DA) (DC)(DG)(DC)(DA)(DC)(DG)(DT) (DA)(DC)(DG)(DC)(DG)(DC)(DT)(DG)(DT)(DC) (DC)(DC)(DC) (DC)(DG)(DC)(DG)(DT)(DT) (DT)(DT)(DA)(DA)(DC)(DC)(DG)(DC)(DC)(DA) (DA)(DG)(DG)(DG) (DG)(DA)(DT)(DT)(DA) (DC)(DA)(DC)(DC)(DC)(DA)(DA)(DG)(DA)(DC) (DA)(DC)(DC)(DA)(DG) (DG)(DC)(DA)(DC) (DG)(DA)(DG)(DA)(DC)(DA)(DG)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DC)(DA) (DA)(DC)(DG) (DA)(DA)(DA)(DA)(DC)(DG)(DG)(DC)(DC)(DA) (DC)(DC)(DA) |
-分子 #6: DNA (153-MER)
| 分子 | 名称: DNA (153-MER) / タイプ: dna / ID: 6 / コピー数: 1 / 分類: DNA |
|---|---|
| 由来(天然) | 生物種: synthetic construct (人工物) |
| 分子量 | 理論値: 47.37107 KDa |
| 配列 | 文字列: (DT)(DG)(DG)(DC)(DC)(DG)(DT)(DT)(DT)(DT) (DC)(DG)(DT)(DT)(DG)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DC)(DT)(DG)(DT)(DC)(DT)(DC) (DG)(DT)(DG)(DC)(DC)(DT)(DG)(DG)(DT)(DG) (DT) (DC)(DT)(DT)(DG)(DG) ...文字列: (DT)(DG)(DG)(DC)(DC)(DG)(DT)(DT)(DT)(DT) (DC)(DG)(DT)(DT)(DG)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DC)(DT)(DG)(DT)(DC)(DT)(DC) (DG)(DT)(DG)(DC)(DC)(DT)(DG)(DG)(DT)(DG) (DT) (DC)(DT)(DT)(DG)(DG)(DG)(DT)(DG) (DT)(DA)(DA)(DT)(DC)(DC)(DC)(DC)(DT)(DT) (DG)(DG) (DC)(DG)(DG)(DT)(DT)(DA)(DA) (DA)(DA)(DC)(DG)(DC)(DG)(DG)(DG)(DG)(DG) (DA)(DC)(DA) (DG)(DC)(DG)(DC)(DG)(DT) (DA)(DC)(DG)(DT)(DG)(DC)(DG)(DT)(DT)(DT) (DA)(DA)(DG)(DC) (DG)(DG)(DT)(DG)(DC) (DT)(DA)(DG)(DA)(DG)(DC)(DT)(DG)(DT)(DC) (DT)(DA)(DC)(DG)(DA) (DC)(DC)(DA)(DA) (DT)(DT)(DG)(DA)(DG)(DC)(DG)(DG)(DC)(DC) (DT)(DC)(DG)(DG)(DC)(DA) (DC)(DC)(DG) (DG)(DG)(DA)(DT)(DT)(DC)(DT)(DG)(DA)(DT) |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 平均電子線量: 58.8 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.5 µm / 最小 デフォーカス(公称値): 1.0 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)