[English] 日本語
 Yorodumi
Yorodumi- EMDB-3809: The Cryo-Electron Microscopy Structure of the Type 1 Chaperone-Us... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3809 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Cryo-Electron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacterial pilus / chaperone-usher pilus / PROTEIN FIBRIL | |||||||||
| Function / homology |  Function and homology information Function and homology informationcell adhesion involved in single-species biofilm formation / pilus / cell adhesion / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Hospenthal MK / Costa TRD / Redzej A / Waksman G | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2017 Journal: Structure / Year: 2017Title: The Cryoelectron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod. Authors: Manuela K Hospenthal / Dawid Zyla / Tiago R D Costa / Adam Redzej / Christoph Giese / James Lillington / Rudi Glockshuber / Gabriel Waksman /   Abstract: Adhesive chaperone-usher pili are long, supramolecular protein fibers displayed on the surface of many bacterial pathogens. The type 1 and P pili of uropathogenic Escherichia coli (UPEC) play ...Adhesive chaperone-usher pili are long, supramolecular protein fibers displayed on the surface of many bacterial pathogens. The type 1 and P pili of uropathogenic Escherichia coli (UPEC) play important roles during urinary tract colonization, mediating attachment to the bladder and kidney, respectively. The biomechanical properties of the helical pilus rods allow them to reversibly uncoil in response to flow-induced forces, allowing UPEC to retain a foothold in the unique and hostile environment of the urinary tract. Here we provide the 4.2-Å resolution cryo-EM structure of the type 1 pilus rod, which together with the previous P pilus rod structure rationalizes the remarkable "spring-like" properties of chaperone-usher pili. The cryo-EM structure of the type 1 pilus rod differs in its helical parameters from the structure determined previously by a hybrid approach. We provide evidence that these structural differences originate from different quaternary structures of pili assembled in vivo and in vitro. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3809.map.gz emd_3809.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3809-v30.xml emd-3809-v30.xml emd-3809.xml emd-3809.xml | 11.8 KB 11.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3809.png emd_3809.png | 273.9 KB | ||
| Masks |  emd_3809_msk_1.map emd_3809_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-3809.cif.gz emd-3809.cif.gz | 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3809 http://ftp.pdbj.org/pub/emdb/structures/EMD-3809 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3809 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3809 | HTTPS FTP |
-Related structure data
| Related structure data |  5oh0MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3809.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3809.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.13 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
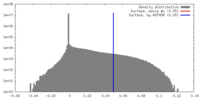
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_3809_msk_1.map emd_3809_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Type 1 Chaperone-usher pilus
| Entire | Name: Type 1 Chaperone-usher pilus |
|---|---|
| Components |
|
-Supramolecule #1: Type 1 Chaperone-usher pilus
| Supramolecule | Name: Type 1 Chaperone-usher pilus / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Superhelical assembly of the pilus rod subunit FimA |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Type-1 fimbrial protein, A chain
| Macromolecule | Name: Type-1 fimbrial protein, A chain / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.835243 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AATTVNGGTV HFKGEVVNAA CAVDAGSVDQ TVQLGQVRTA SLAQEGATSS AVGFNIQLND CDTNVASKAA VAFLGTAIDA GHTNVLALQ SSAAGSATNV GVQILDRTGA ALTLDGATFS SETTLNNGTN TIPFQARYFA TGAATPGAAN ADATFKVQYQ UniProtKB: Type-1 fimbrial protein, A chain |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Component:
| ||||||
|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil 1.2/1.3 400 mesh grid / Pretreatment - Type: GLOW DISCHARGE | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 8.01188 Å Applied symmetry - Helical parameters - Δ&Phi: 114.992 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 4.2 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 2.0) / Number images used: 115510 |
|---|---|
| Segment selection | Number selected: 115545 / Software - Name: RELION (ver. 2.0) |
| Startup model | Type of model: OTHER / Details: solid cylinder (diameter 100 Angstrom) |
| Final angle assignment | Type: NOT APPLICABLE |
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Output model |  PDB-5oh0: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)