+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Partially closed falcilysin, from free falcilysin dataset | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | falcilysin partially closed conformation / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationhemoglobin catabolic process / apicoplast / food vacuole / Hydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases / vacuolar membrane / protein processing / metalloendopeptidase activity / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
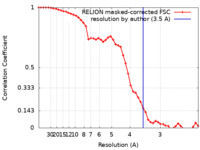
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Lin JQ / Yan XF / Lescar J | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2024 Journal: Commun Biol / Year: 2024Title: Inhibition of falcilysin from Plasmodium falciparum by interference with its closed-to-open dynamic transition. Authors: Jianqing Lin / Xinfu Yan / Zara Chung / Chong Wai Liew / Abbas El Sahili / Evgeniya V Pechnikova / Peter R Preiser / Zbynek Bozdech / Yong-Gui Gao / Julien Lescar /   Abstract: In the absence of an efficacious vaccine, chemotherapy remains crucial to prevent and treat malaria. Given its key role in haemoglobin degradation, falcilysin constitutes an attractive target. Here, ...In the absence of an efficacious vaccine, chemotherapy remains crucial to prevent and treat malaria. Given its key role in haemoglobin degradation, falcilysin constitutes an attractive target. Here, we reveal the mechanism of enzymatic inhibition of falcilysin by MK-4815, an investigational new drug with potent antimalarial activity. Using X-ray crystallography, we determine two binary complexes of falcilysin in a closed state, bound with peptide substrates from the haemoglobin α and β chains respectively. An antiparallel β-sheet is formed between the substrate and enzyme, accounting for sequence-independent recognition at positions P2 and P1. In contrast, numerous contacts favor tyrosine and phenylalanine at the P1' position of the substrate. Cryo-EM studies reveal a majority of unbound falcilysin molecules adopting an open conformation. Addition of MK-4815 shifts about two-thirds of falcilysin molecules to a closed state. These structures give atomic level pictures of the proteolytic cycle, in which falcilysin interconverts between a closed state conducive to proteolysis, and an open conformation amenable to substrate diffusion and products release. MK-4815 and quinolines bind to an allosteric pocket next to a hinge region of falcilysin and hinders this dynamic transition. These data should inform the design of potent inhibitors of falcilysin to combat malaria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37940.map.gz emd_37940.map.gz | 6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37940-v30.xml emd-37940-v30.xml emd-37940.xml emd-37940.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37940_fsc.xml emd_37940_fsc.xml | 4.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_37940.png emd_37940.png | 20.4 KB | ||
| Masks |  emd_37940_msk_1.map emd_37940_msk_1.map | 8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-37940.cif.gz emd-37940.cif.gz | 6.1 KB | ||
| Others |  emd_37940_additional_1.map.gz emd_37940_additional_1.map.gz emd_37940_additional_2.map.gz emd_37940_additional_2.map.gz emd_37940_half_map_1.map.gz emd_37940_half_map_1.map.gz emd_37940_half_map_2.map.gz emd_37940_half_map_2.map.gz | 1.2 MB 7.5 MB 6 MB 6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37940 http://ftp.pdbj.org/pub/emdb/structures/EMD-37940 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37940 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37940 | HTTPS FTP |
-Related structure data
| Related structure data |  8wyxMC  8wxwC  8wxzC  8wytC  8wyuC  8wyyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37940.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37940.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.146 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_37940_msk_1.map emd_37940_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #2
| File | emd_37940_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_37940_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_37940_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_37940_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Falcilysin
| Entire | Name: Falcilysin |
|---|---|
| Components |
|
-Supramolecule #1: Falcilysin
| Supramolecule | Name: Falcilysin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Falcilysin
| Macromolecule | Name: Falcilysin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 132.220375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EWIHEKSPKH NSYDIIEKRY NEEFKMTYTV YQHKKAKTQV ISLGTNDPLD VEQAFAFYVK TLTHSGKGIP HILEHSVLSG SKNYNYKNS IGLLEKGTLH THLNAYTFND RTVYMAGSMN NKDFFNIMGV YMDSVFQPNV LENKYIFETE GWTYEVEKLK E DEKGKAEI ...String: EWIHEKSPKH NSYDIIEKRY NEEFKMTYTV YQHKKAKTQV ISLGTNDPLD VEQAFAFYVK TLTHSGKGIP HILEHSVLSG SKNYNYKNS IGLLEKGTLH THLNAYTFND RTVYMAGSMN NKDFFNIMGV YMDSVFQPNV LENKYIFETE GWTYEVEKLK E DEKGKAEI PQMKDYKVSF NGIVYNEMKG ALSSPLEDLY HEEMKYMFPD NVHSNNSGGD PKEITNLTYE EFKEFYYKNY NP KKVKVFF FSKNNPTELL NFVDQYLGQL DYSKYRDDAV ESVEYQTYKK GPFYIKKKYG DHSEEKENLV SVAWLLNPKV DKT NNHNNN HSNNQSSENN GYSNGSHSSD LSLENPTDYF VLLIINNLLI HTPESVLYKA LTDCGLGNNV IDRGLNDSLV QYIF SIGLK GIKRNNEKIK NFDKVHYEVE DVIMNALKKV VKEGFNKSAV EASINNIEFI LKEANLKTSK SIDFVFEMTS KLNYN RDPL LIFEFEKYLN IVKNKIKNEP MYLEKFVEKH FINNAHRSVI LLEGDENYAQ EQENLEKQEL KKRIENFNEQ EKEQVI KNF EELSKYKNAE ESPEHLNKFP IISISDLNKK TLEVPVNVYF TNINENNNIM ETYNKLKTNE HMLKDNMDVF LKKYVLK ND KHNTNNNNNN NNNMDYSFTE TKYEGNVPIL VYEMPTTGIV YLQFVFSLDH LTVDELAYLN LFKTLILENK TNKRSSED F VILREKNIGS MSANVALYSK DDHLNVTDKY NAQALFNLEM HVLSHKCNDA LNIALEAVKE SDFSNKKKVI DILKRKING MKTTFSEKGY AILMKYVKAH LNSKHYAHNI IYGYENYLKL QEQLELAEND FKTLENILVR IRNKIFNKKN LMVSVTSDYG ALKHLFVNS NESLKNLVSY FEENDKYIND MQNKVNDPTV MGWNEEIKSK KLFDEEKVKK EFFVLPTFVN SVSMSGILFK P GEYLDPSF TVIVAALKNS YLWDTVRGLN GAYGVFADIE YDGSVVFLSA RDPNLEKTLA TFRESAKGLR KMADTMTEND LL RYIINTI GTIDKPRRGI ELSKLSFLRL ISNESEQDRV EFRKRIMNTK KEDFYKFADL LESKVNEFEK NIVIITTKEK ANE YIANVD GEFKKVLI UniProtKB: Falcilysin |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20 mM Na HEPES, 300 mM NaCl, 0.5 mM TCEP, pH 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)