[English] 日本語
 Yorodumi
Yorodumi- EMDB-37253: Structure of Tomato spotted wilt virus L protein contained endoH ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Tomato spotted wilt virus L protein contained endoH domain binding to 3'5'vRNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Tomato spotted wilt virus / L protein / VIRAL PROTEIN / VIRAL PROTEIN-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA-directed RNA polymerase / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription Similarity search - Function | |||||||||
| Biological species |  Tomato spotted wilt virus (strain Bulgarian L3) / synthetic construct (others) Tomato spotted wilt virus (strain Bulgarian L3) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Cao L / Wang X | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2025 Journal: Nat Plants / Year: 2025Title: Structural basis for the activation of plant bunyavirus replication machinery and its dual-targeted inhibition by ribavirin. Authors: Jia Li / Lei Cao / Yaqian Zhao / Jinghan Shen / Lei Wang / Mingfeng Feng / Min Zhu / Yonghao Ye / Richard Kormelink / Xiaorong Tao / Xiangxi Wang /   Abstract: Despite the discovery of plant viruses as a new class of pathogens over a century ago, the structure of plant virus replication machinery and antiviral pesticide remains lacking. Here we report five ...Despite the discovery of plant viruses as a new class of pathogens over a century ago, the structure of plant virus replication machinery and antiviral pesticide remains lacking. Here we report five cryogenic electron microscopy structures of a ~330-kDa RNA-dependent RNA polymerase (RdRp) from a devastating plant bunyavirus, tomato spotted wilt orthotospovirus (TSWV), including the apo, viral-RNA-bound, base analogue ribavirin-bound and ribavirin-triphosphate-bound states. They reveal that a flexible loop of RdRp's motif F functions as 'sensor' to perceive viral RNA and further acts as an 'adaptor' to promote the formation of a complete catalytic centre. A ten-base RNA 'hook' structure is sufficient to trigger major conformational changes and activate RdRp. Chemical screening showed that ribavirin is effective against TSWV, and structural data revealed that ribavirin disrupts both hook-binding and catalytic core formation, locking polymerase in its inactive state. This work provides structural insights into the mechanisms of plant bunyavirus RdRp activation and its dual-targeted site inhibition, facilitating the development of pesticides against plant viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37253.map.gz emd_37253.map.gz | 38.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37253-v30.xml emd-37253-v30.xml emd-37253.xml emd-37253.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37253.png emd_37253.png | 105.5 KB | ||
| Filedesc metadata |  emd-37253.cif.gz emd-37253.cif.gz | 7.6 KB | ||
| Others |  emd_37253_half_map_1.map.gz emd_37253_half_map_1.map.gz emd_37253_half_map_2.map.gz emd_37253_half_map_2.map.gz | 37.8 MB 37.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37253 http://ftp.pdbj.org/pub/emdb/structures/EMD-37253 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37253 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37253 | HTTPS FTP |
-Validation report
| Summary document |  emd_37253_validation.pdf.gz emd_37253_validation.pdf.gz | 902.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37253_full_validation.pdf.gz emd_37253_full_validation.pdf.gz | 901.9 KB | Display | |
| Data in XML |  emd_37253_validation.xml.gz emd_37253_validation.xml.gz | 11.4 KB | Display | |
| Data in CIF |  emd_37253_validation.cif.gz emd_37253_validation.cif.gz | 13.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37253 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37253 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37253 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37253 | HTTPS FTP |
-Related structure data
| Related structure data |  8ki7MC  8ki6C  8ki8C  8ki9C  8kiaC  9j8vC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_37253.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37253.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
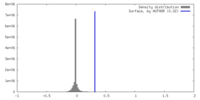
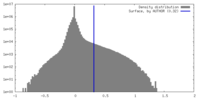
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Structure of Tomato spotted wilt virus L protein contained endoH ...
| Entire | Name: Structure of Tomato spotted wilt virus L protein contained endoH domain binding to 3'5'vRNA |
|---|---|
| Components |
|
-Supramolecule #1: Structure of Tomato spotted wilt virus L protein contained endoH ...
| Supramolecule | Name: Structure of Tomato spotted wilt virus L protein contained endoH domain binding to 3'5'vRNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Tomato spotted wilt virus (strain Bulgarian L3) Tomato spotted wilt virus (strain Bulgarian L3) |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Tomato spotted wilt virus (strain Bulgarian L3) / Strain: Bulgarian L3 Tomato spotted wilt virus (strain Bulgarian L3) / Strain: Bulgarian L3 |
| Molecular weight | Theoretical: 240.378375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNIQKIQKLI ENGTTLLLSI EDCVGSNYDL ALDLHKRNSD EIPEDVIINN NAKNYETMRE LIVKITADGE GLNKGMATVD VKKLSEMVS LFEQKYLETE LARHDIFGEL ISRHLRIKPK QRSEVEIEHA LREYLDELNK KSCINKLSDD EFERINKEYV A TNATPDNY ...String: MNIQKIQKLI ENGTTLLLSI EDCVGSNYDL ALDLHKRNSD EIPEDVIINN NAKNYETMRE LIVKITADGE GLNKGMATVD VKKLSEMVS LFEQKYLETE LARHDIFGEL ISRHLRIKPK QRSEVEIEHA LREYLDELNK KSCINKLSDD EFERINKEYV A TNATPDNY VIYKESKNSE LCLIIYDWKI SVDARTETKT MEKYYKNIWK SFKDIKVNGK PFLEDHPVFV SIVILKPIAG MP ITVTSSR VLEKFEDSPS ALHGERIKHA RNAKLLNISH VGQIVGTTPT VVRNYYANTQ KIKSEVRGIL GDDFGSKDVF FSH WTSKYK ERNPTEIAYS EDIERIIDSL VTDEITKEEI IHFLFGNFCF HIETMNDQHI ADKFKGYQSS CINLKIEPKV DLAD LKDHL IQKQQIWESL YGKHLEKIML RIREKKKKEK EIPDITTAFN QNAAEYEEKY PNCFTNDLSE TKTNFSMTWS PSFEK IELS SEVDYNNAII NKFRESFKSS SRVIYNSPYS SINNQTNKAR DITNLVRLCL TELSCDTTKM EKQELEDEID INTGSI KVE RTKKSKEWNK QGSCLTRNKN EFCMKETGRE NKTIYFKGLA VMNIGMSSKK RILKKEEIKE RISKGLEYDT SERQADP ND DYSSIDMSSL THMKKLIRHD NEDSLSWCER IKDSLFVLHN GDIREEGKIT SVYNNYAKNP ECLYIQDSVL KTELETCK K INKLCNDLAI YHYSEDMMQF SKGLMVADRY MTKESFKILT TANTSMMLLA FKGDGMNTGG SGVPYIALHI VDEDMSDQF NICYTKEIYS YFRNGSNYIY IMRPQRLNQV RLLSLFKTPS KVPVCFAQFS KKANEMEKWL KNKDIEKVNV FSMTMTVKQI LINIVFSSV MIGTVTKLSR MGIFDFMRYA GFLPLSDYSN IKEYIRDKFD PDITNVADIY FVNGIKKLLF RMEDLNLSTN A KPVVVDHE NDIIGGITDL NIKCPITGST LLTLEDLYNN VYLAIYMMPK SLHNHVHNLT SLLNVPAEWE LKFRKELGFN IF EDIYPKK AMFDDKDLFS INGALNVKAL SDYYLGNIEN VGLMRSEIEN KEDFLSPCYK ISTLKSSKKC SQSNIISTDE IIE CLQNAK IQDIENWKGN NLAIIKGLIR TYNEEKNRLV EFFEDNCVNS LYLVEKLKEI INSGSITVGK SVTSKFIRNN HPLT VETYL KTKLYYRNNV TVLKSKKVSE ELYDLVKQFH NMMEIDLDSV MNLGKGTEGK KHTFLQMLEF VMSKAKNVTG SVDFL VSVF EKMQRTKTDR EIYLMSMKVK MMLYFIEHTF KHVAQSDPSE AISISGDNKI RALSTLSLDT ITSYNDILNK NSKKSR LAF LSADQSKWSA SDLTYKYVLA IILNPILTTG EASLMIECIL MYVKLKKVCI PTDIFLNLRK AQGTFGQNET AIGLLTK GL TTNTYPVSMN WLQGNLNYLS SVYHSCAMKA YHKTLECYKD CDFQTRWIVH SDDNATSLIA SGEVDKMLTD FSSSSLPE M LFRSIEAHFK SFCITLNPKK SYASSSEVEF ISERIVNGAI IPLYCRHLAN CCTESSHISY FDDLMSLSIH VTMLLRKGC PNEVIPFAYG AVQVQALSIY SMLPGEVNDS IRIFKKLGVS LKSNEIPTNM GGWLTSPIEP LSILGPSSND QIIYYNVIRD FLNKKSLEE VKDSVSSSSY LQMRFRELKG KYEKGTLEEK DKKMIFLINL FEKASVSEDS DVLTIGMKFQ TMLTQIIKLP N FINENALN KMSSYKDFSK LYPNLKKNED LYKSTKNLKI DEDAILEEDE LYEKIASSLE MESVHDIMIK NPETILIAPL ND RDFLLSQ LFMYTSPSKR NQLSNQSTEK LALDRVLRSK ARTFVDISST VKMTYEENME KKILEMLKFD LDSYCSFKTC VNL VIKDVN FSMLIPILDS AYPCESRKRD NYNFRWFQTE KWIPVVEGSP GLVVMHAVYG SNYIENLGLK NIPLTDDSIN VLTS TFGTG LIMEDVKSLV KGKDSFETEA FSNSNECQRL VKACNYMIAA QNRLLAINTC FTRKSFPFYS KFNLGRGFIS NTLAL LSTI YSKEES UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: RNA (5'-R(P*AP*GP*AP*GP*CP*AP*AP*UP*CP*A)-3')
| Macromolecule | Name: RNA (5'-R(P*AP*GP*AP*GP*CP*AP*AP*UP*CP*A)-3') / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 3.208012 KDa |
| Sequence | String: AGAGCAAUCA |
-Macromolecule #3: RNA (5'-R(P*GP*CP*AP*AP*UP*CP*AP*GP*G)-3')
| Macromolecule | Name: RNA (5'-R(P*GP*CP*AP*AP*UP*CP*AP*GP*G)-3') / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 2.894807 KDa |
| Sequence | String: GCAAUCAGG |
-Macromolecule #4: RNA (5'-R(P*AP*CP*CP*UP*GP*AP*UP*UP*GP*CP*UP*CP*U)-3')
| Macromolecule | Name: RNA (5'-R(P*AP*CP*CP*UP*GP*AP*UP*UP*GP*CP*UP*CP*U)-3') type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.055421 KDa |
| Sequence | String: ACCUGAUUGC UCU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)