[English] 日本語
 Yorodumi
Yorodumi- EMDB-36441: Cryo-EM structure of dengue virus serotype 3 strain CH53489 spher... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of dengue virus serotype 3 strain CH53489 spherical particle in complex with human antibody DENV-290 Fab at 37 deg C | |||||||||
 Map data Map data | postprocessed map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | dengue virus / human antibody / dengue-antibody structure / virus | |||||||||
| Biological species |  Dengue virus type 3 / Dengue virus type 3 /  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.6 Å | |||||||||
 Authors Authors | Fibriansah G / Ng TS / Tan AWK / Shi J / Lok SM | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Ultrapotent human antibodies lock E protein dimers central region of diverse DENV3 morphological variants. Authors: Guntur Fibriansah / Thiam-Seng Ng / Xin-Ni Lim / Anastasia Shebanova / Lee Ching Ng / Song Ling Tan / Aaron W K Tan / Jian Shi / James E Crowe / Shee-Mei Lok /   Abstract: Dengue virus (DENV) consists of four serotypes (DENV1-4). Current vaccines induce differing levels of immune response against the four serotypes, that might prime recipients to develop severe disease ...Dengue virus (DENV) consists of four serotypes (DENV1-4). Current vaccines induce differing levels of immune response against the four serotypes, that might prime recipients to develop severe disease in subsequent infections. Several DENV tetravalent vaccine clinical trials suggested an increased incidence in severe DENV3 cases, suggesting a need to develop DENV3 therapeutics. Human monoclonal antibodies (HMAbs) DENV-290 and DENV-115 are ultrapotent against diverse DENV3 strains with differing particle morphologies. They mainly neutralize by inhibition of virus attachment to cells. CryoEM structures of Fabs complexed with differing DENV3 morphological variants show their Fabs binding across two E protein protomers at the center of the E dimer. This new class of E protein dimer binding antibodies is named EDE-C. The cryoEM structures also show how IgGs engage the DENV particles. Results define the structural and molecular basis for the ultrapotent activity of EDE-C antibodies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36441.map.gz emd_36441.map.gz | 472.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36441-v30.xml emd-36441-v30.xml emd-36441.xml emd-36441.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36441.png emd_36441.png | 232.5 KB | ||
| Masks |  emd_36441_msk_1.map emd_36441_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36441.cif.gz emd-36441.cif.gz | 6 KB | ||
| Others |  emd_36441_half_map_1.map.gz emd_36441_half_map_1.map.gz emd_36441_half_map_2.map.gz emd_36441_half_map_2.map.gz | 404.2 MB 404.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36441 http://ftp.pdbj.org/pub/emdb/structures/EMD-36441 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36441 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36441 | HTTPS FTP |
-Validation report
| Summary document |  emd_36441_validation.pdf.gz emd_36441_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36441_full_validation.pdf.gz emd_36441_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_36441_validation.xml.gz emd_36441_validation.xml.gz | 19.1 KB | Display | |
| Data in CIF |  emd_36441_validation.cif.gz emd_36441_validation.cif.gz | 23 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36441 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36441 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36441 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36441 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36441.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36441.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocessed map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.71 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36441_msk_1.map emd_36441_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
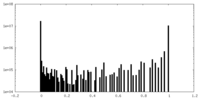
| Density Histograms |
-Half map: half map 1
| File | emd_36441_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
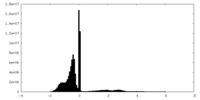
| Density Histograms |
-Half map: half map 2
| File | emd_36441_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
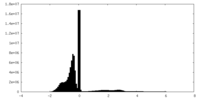
| Density Histograms |
- Sample components
Sample components
-Entire : Dengue virus serotype 3 strain CH53489 in complex with human anti...
| Entire | Name: Dengue virus serotype 3 strain CH53489 in complex with human antibody DENV-290 Fab at 37 deg C |
|---|---|
| Components |
|
-Supramolecule #1: Dengue virus serotype 3 strain CH53489 in complex with human anti...
| Supramolecule | Name: Dengue virus serotype 3 strain CH53489 in complex with human antibody DENV-290 Fab at 37 deg C type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Dengue virus serotype 3 strain CH53489
| Supramolecule | Name: Dengue virus serotype 3 strain CH53489 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Dengue virus type 3 Dengue virus type 3 |
-Supramolecule #3: Human antibody DENV-290 Fab (heavy and light chain)
| Supramolecule | Name: Human antibody DENV-290 Fab (heavy and light chain) / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Envelope protein
| Macromolecule | Name: Envelope protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: mrcvgvgnrd fveglsgat w vdvvlehg gc vttmakn kpt ldielq ktea tqlat lrklc iegk itnitt dsr cptqgea vl peeqdqny v ckhtyvdrg wgngcglfgk gslvtcakf q clepiegk vv qyenlky tvi itvhtg dqhq vgnet ...String: mrcvgvgnrd fveglsgat w vdvvlehg gc vttmakn kpt ldielq ktea tqlat lrklc iegk itnitt dsr cptqgea vl peeqdqny v ckhtyvdrg wgngcglfgk gslvtcakf q clepiegk vv qyenlky tvi itvhtg dqhq vgnet qgvta eitp qastte ail peygtlg le csprtgld f nemilltmk nkawmvhrqw ffdlplpwa s gattetpt wn rkellvt fkn ahakkq evvv lgsqe gamht altg ateiqn sgg tsifagh lk crlkmdkl e lkgmsyamc tntfvlkkev setqhgtil i kveykged ap ckipfst edg qgkahn grli tanpv vtkke epvn ieaepp fge snivigi gd nalkinwy k kgssigkmf eatargarrm ailgdtawd f gsvggvln sl gkmvhqi fgs aytalf sgvs wvmki gigvl ltwi glnskn tsm sfsciai gi itlylgav v qa |
-Macromolecule #2: Membrane protein
| Macromolecule | Name: Membrane protein / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: svala phvg mgldtr tqt wmsaega wr qvekvetw a lrhpgftil alflahyigt sltqkvvif i llmlvtps mt |
-Macromolecule #3: Human antibody DENV-290 heavy chain
| Macromolecule | Name: Human antibody DENV-290 heavy chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVELVESGGD VVQPGKSLRL SCAASGFTFT NYAMHWLRQA PGKGLEWVAV ISSDVNDKYY ADSVKGRFTI SRDNSKNTLY LQMNSLTPED TAVYYCAREQ AVGTNPWAFD YWGQGTLVTV SS |
-Macromolecule #4: Human antibody DENV-290 light chain
| Macromolecule | Name: Human antibody DENV-290 light chain / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: HIVMTQSPLS LSVTPGQPAS ISCKSSQISS WGSDGKTYLY WYLQKPGQSP QLLIYEVSSR FSGVSDRFSG SGSGTDFTLK ISRVQAEDVG LYYCMQGLHL PLTFGQGTRL EIK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Details: NTE buffer (12 mM Tris-HCl pH 8.0, 120 mM NaCl and 1 mM EDTA) |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Details | The purified virus was mixed with DENV-290 Fab at a molar ratio of 1.5 Fab for every E-protein and then the mixture was incubated at 4 deg C for 30 min. The complex was further incubated at 37 deg C for 30 min, and then incubated at 4 deg C for another 2 h |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 18.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)