[English] 日本語
 Yorodumi
Yorodumi- EMDB-36436: Cryo-EM structure of dengue virus serotype 3 strain EHIE46200Y19 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of dengue virus serotype 3 strain EHIE46200Y19 in complex with human antibody DENV-115 IgG at 4 deg C | |||||||||
 Map data Map data | postprocessed map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | dengue virus / human antibody / dengue-antibody structure / virus | |||||||||
| Biological species |  Dengue virus type 3 / Dengue virus type 3 /  Homo sapiens (human) / Homo sapiens (human) /  dengue virus type 3 dengue virus type 3 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Fibriansah G / Ng TS / Tan AWK / Shi J / Lok SM | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Ultrapotent human antibodies lock E protein dimers central region of diverse DENV3 morphological variants. Authors: Guntur Fibriansah / Thiam-Seng Ng / Xin-Ni Lim / Anastasia Shebanova / Lee Ching Ng / Song Ling Tan / Aaron W K Tan / Jian Shi / James E Crowe / Shee-Mei Lok /   Abstract: Dengue virus (DENV) consists of four serotypes (DENV1-4). Current vaccines induce differing levels of immune response against the four serotypes, that might prime recipients to develop severe disease ...Dengue virus (DENV) consists of four serotypes (DENV1-4). Current vaccines induce differing levels of immune response against the four serotypes, that might prime recipients to develop severe disease in subsequent infections. Several DENV tetravalent vaccine clinical trials suggested an increased incidence in severe DENV3 cases, suggesting a need to develop DENV3 therapeutics. Human monoclonal antibodies (HMAbs) DENV-290 and DENV-115 are ultrapotent against diverse DENV3 strains with differing particle morphologies. They mainly neutralize by inhibition of virus attachment to cells. CryoEM structures of Fabs complexed with differing DENV3 morphological variants show their Fabs binding across two E protein protomers at the center of the E dimer. This new class of E protein dimer binding antibodies is named EDE-C. The cryoEM structures also show how IgGs engage the DENV particles. Results define the structural and molecular basis for the ultrapotent activity of EDE-C antibodies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36436.map.gz emd_36436.map.gz | 459.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36436-v30.xml emd-36436-v30.xml emd-36436.xml emd-36436.xml | 22.2 KB 22.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36436.png emd_36436.png | 263.2 KB | ||
| Masks |  emd_36436_msk_1.map emd_36436_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36436.cif.gz emd-36436.cif.gz | 5.6 KB | ||
| Others |  emd_36436_half_map_1.map.gz emd_36436_half_map_1.map.gz emd_36436_half_map_2.map.gz emd_36436_half_map_2.map.gz | 398.4 MB 398.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36436 http://ftp.pdbj.org/pub/emdb/structures/EMD-36436 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36436 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36436 | HTTPS FTP |
-Validation report
| Summary document |  emd_36436_validation.pdf.gz emd_36436_validation.pdf.gz | 980.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36436_full_validation.pdf.gz emd_36436_full_validation.pdf.gz | 980.5 KB | Display | |
| Data in XML |  emd_36436_validation.xml.gz emd_36436_validation.xml.gz | 19.1 KB | Display | |
| Data in CIF |  emd_36436_validation.cif.gz emd_36436_validation.cif.gz | 23 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36436 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36436 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36436 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36436 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36436.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36436.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocessed map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.622 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36436_msk_1.map emd_36436_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
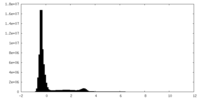
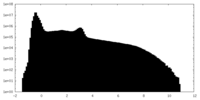
| Density Histograms |
-Half map: half map 1
| File | emd_36436_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
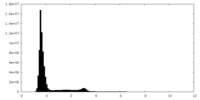
| Density Histograms |
-Half map: half map 2
| File | emd_36436_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
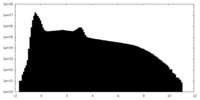
| Density Histograms |
- Sample components
Sample components
-Entire : Dengue virus serotype 3 strain EHIE46200Y19 in complex with human...
| Entire | Name: Dengue virus serotype 3 strain EHIE46200Y19 in complex with human antibody DENV-115 IgG at 4 deg C |
|---|---|
| Components |
|
-Supramolecule #1: Dengue virus serotype 3 strain EHIE46200Y19 in complex with human...
| Supramolecule | Name: Dengue virus serotype 3 strain EHIE46200Y19 in complex with human antibody DENV-115 IgG at 4 deg C type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Dengue virus serotype 3 strain EHIE46200Y19
| Supramolecule | Name: Dengue virus serotype 3 strain EHIE46200Y19 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Dengue virus type 3 Dengue virus type 3 |
-Supramolecule #3: Human antibody DENV-115 IgG (heavy and light chain)
| Supramolecule | Name: Human antibody DENV-115 IgG (heavy and light chain) / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Envelope protein
| Macromolecule | Name: Envelope protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  dengue virus type 3 dengue virus type 3 |
| Sequence | String: MRCVGVGNRD FVEGLSGATW VDVVLEHGGC VTTMAKNKPT LDIELQKTEA TQLATLRKLC IEGKITNVTT DSRCPTQGEA ILPEEQDQN YVCKHTYVDR GWGNGCGLFG KGSLVTCAKF QCLELIEGKV VQHENLKYTV IITVHTGDQH QVGNETQGVT A EITPQAST ...String: MRCVGVGNRD FVEGLSGATW VDVVLEHGGC VTTMAKNKPT LDIELQKTEA TQLATLRKLC IEGKITNVTT DSRCPTQGEA ILPEEQDQN YVCKHTYVDR GWGNGCGLFG KGSLVTCAKF QCLELIEGKV VQHENLKYTV IITVHTGDQH QVGNETQGVT A EITPQAST VEAILPEYGT LGLECSPRTG LDFNEMILLT MKNKAWMVHR QWFFDLPLPW TSGATTETPT WNKKELLVTF KN AHAKKQE VVVLGSQEGA MHTALTGATE IQTSGGTSIF AGHLKCRLKM DKLELKGMSY AMCSNAFVLK KEVSETQHGT ILI KVEYKG EDAPCKIPFS TEDGQGKAHN GRLITANPVV TKKEELVNIE AEPPFGESNI IIGIGDKALR INWYKKGSSI GKMF EATAR GARRMAILGD TAWDFGSVGG VLNSLGKMVH QIFGSAYTAL FSGVSWIMKI GIGVLLTWIG LNSKNTSMSF SCIVI GIIT LYLGAVVQA |
-Macromolecule #2: Membrane protein
| Macromolecule | Name: Membrane protein / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  dengue virus type 3 dengue virus type 3 |
| Sequence | String: SVALAPHVGM GLDTRAQTWM SAEGAWRQIE KVETWAFRHP GFTILALFLA HYIGTSLTQK VVIFILLMLV TPSMT |
-Macromolecule #3: Human antibody DENV-115 heavy chain
| Macromolecule | Name: Human antibody DENV-115 heavy chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGAPVKV SCEASGYTFT DYFIHWVRQA PGQGLEWMGW INPISGGTNY HPRFHGGVTM TRDTSMKVAY MELKRLTSD DTAVYFCARG RDFRGGYSQL DYWGQGTLVT VSS |
-Macromolecule #4: Human antibody DENV-115 light chain
| Macromolecule | Name: Human antibody DENV-115 light chain / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVLTQPPSA SGTPGQRVTI SCSGGSSNIA INTVNWYQQV PGTAPKLLMY SNNQRPSGVP DRFSGSKSGT SASLAISGLQ SEDEADYYC ATWDDSLKDV LFGGGTKLTV L |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Details: NTE buffer (12 mM Tris-HCl pH 8.0, 120 mM NaCl and 1 mM EDTA) |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Details | The purified virus was mixed with DENV-115 IgG at a molar ratio of 1.5 Fab for every E-protein and then the mixture was incubated at 4 deg C for 30 min. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 22.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.6000000000000005 µm / Nominal defocus min: 0.4 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)