+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Dopamine Receptor D5R-Gs-Rotigotine complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Dopamine / Dopamine receptor / GPCR / D5R / Gs / Rotigotine / Parkinson's disease / Restless legs syndrome / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information: / regulation of female receptivity / norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure / dopamine neurotransmitter receptor activity, coupled via Gs / dopamine neurotransmitter receptor activity / positive regulation of adenylate cyclase activity / regulation of systemic arterial blood pressure by vasopressin / Dopamine receptors / dopamine binding / phospholipase C-activating dopamine receptor signaling pathway ...: / regulation of female receptivity / norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure / dopamine neurotransmitter receptor activity, coupled via Gs / dopamine neurotransmitter receptor activity / positive regulation of adenylate cyclase activity / regulation of systemic arterial blood pressure by vasopressin / Dopamine receptors / dopamine binding / phospholipase C-activating dopamine receptor signaling pathway / sensitization / synaptic transmission, dopaminergic / non-motile cilium / long-term synaptic depression / ciliary membrane / G protein-coupled dopamine receptor signaling pathway / transmission of nerve impulse / associative learning / activation of adenylate cyclase activity / negative regulation of blood pressure / adenylate cyclase-activating adrenergic receptor signaling pathway / reactive oxygen species metabolic process / response to amphetamine / response to cocaine / wound healing / brush border membrane / electron transport chain / G protein-coupled receptor activity / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / intracellular calcium ion homeostasis / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / chemical synaptic transmission / Ras protein signal transduction / electron transfer activity / periplasmic space / Extra-nuclear estrogen signaling / cell population proliferation / positive regulation of MAPK cascade / cilium / iron ion binding / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / heme binding / synapse / protein-containing complex binding / signal transduction / extracellular exosome / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Xu P / Huang S / Zhuang Y / Mao C / Zhang Y / Wang Y / Li H / Jiang Y / Xu HE | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2023 Journal: Cell Res / Year: 2023Title: Structural genomics of the human dopamine receptor system. Authors: Peiyu Xu / Sijie Huang / Brian E Krumm / Youwen Zhuang / Chunyou Mao / Yumu Zhang / Yue Wang / Xi-Ping Huang / Yong-Feng Liu / Xinheng He / Huadong Li / Wanchao Yin / Yi Jiang / Yan Zhang / ...Authors: Peiyu Xu / Sijie Huang / Brian E Krumm / Youwen Zhuang / Chunyou Mao / Yumu Zhang / Yue Wang / Xi-Ping Huang / Yong-Feng Liu / Xinheng He / Huadong Li / Wanchao Yin / Yi Jiang / Yan Zhang / Bryan L Roth / H Eric Xu /   Abstract: The dopaminergic system, including five dopamine receptors (D1R to D5R), plays essential roles in the central nervous system (CNS); and ligands that activate dopamine receptors have been used to ...The dopaminergic system, including five dopamine receptors (D1R to D5R), plays essential roles in the central nervous system (CNS); and ligands that activate dopamine receptors have been used to treat many neuropsychiatric disorders, including Parkinson's Disease (PD) and schizophrenia. Here, we report cryo-EM structures of all five subtypes of human dopamine receptors in complex with G protein and bound to the pan-agonist, rotigotine, which is used to treat PD and restless legs syndrome. The structures reveal the basis of rotigotine recognition in different dopamine receptors. Structural analysis together with functional assays illuminate determinants of ligand polypharmacology and selectivity. The structures also uncover the mechanisms of dopamine receptor activation, unique structural features among the five receptor subtypes, and the basis of G protein coupling specificity. Our work provides a comprehensive set of structural templates for the rational design of specific ligands to treat CNS diseases targeting the dopaminergic system. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35687.map.gz emd_35687.map.gz | 28.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35687-v30.xml emd-35687-v30.xml emd-35687.xml emd-35687.xml | 23.2 KB 23.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35687.png emd_35687.png | 65.6 KB | ||
| Filedesc metadata |  emd-35687.cif.gz emd-35687.cif.gz | 7.4 KB | ||
| Others |  emd_35687_half_map_1.map.gz emd_35687_half_map_1.map.gz emd_35687_half_map_2.map.gz emd_35687_half_map_2.map.gz | 23.2 MB 23.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35687 http://ftp.pdbj.org/pub/emdb/structures/EMD-35687 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35687 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35687 | HTTPS FTP |
-Validation report
| Summary document |  emd_35687_validation.pdf.gz emd_35687_validation.pdf.gz | 722.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35687_full_validation.pdf.gz emd_35687_full_validation.pdf.gz | 721.9 KB | Display | |
| Data in XML |  emd_35687_validation.xml.gz emd_35687_validation.xml.gz | 10.5 KB | Display | |
| Data in CIF |  emd_35687_validation.cif.gz emd_35687_validation.cif.gz | 12.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35687 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35687 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35687 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35687 | HTTPS FTP |
-Related structure data
| Related structure data |  8irvMC  8irrC  8irsC  8irtC  8iruC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35687.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35687.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.071 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35687_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_35687_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dopamine Receptor D5R-Gs-Rotigotine complex
| Entire | Name: Dopamine Receptor D5R-Gs-Rotigotine complex |
|---|---|
| Components |
|
-Supramolecule #1: Dopamine Receptor D5R-Gs-Rotigotine complex
| Supramolecule | Name: Dopamine Receptor D5R-Gs-Rotigotine complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.74827 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GCTLSAEDKA AVERSKMIEK QLQKDKQVYR ATHRLLLLGA DNSGKSTIVK QMRIYHVNGY SEEECKQYKA VVYSNTIQSI IAIIRAMGR LKIDFGDSAR ADDARQLFVL AGAAEEGFMT AELAGVIKRL WKDSGVQACF NRSREYQLND SAAYYLNDLD R IAQPNYIP ...String: GCTLSAEDKA AVERSKMIEK QLQKDKQVYR ATHRLLLLGA DNSGKSTIVK QMRIYHVNGY SEEECKQYKA VVYSNTIQSI IAIIRAMGR LKIDFGDSAR ADDARQLFVL AGAAEEGFMT AELAGVIKRL WKDSGVQACF NRSREYQLND SAAYYLNDLD R IAQPNYIP TQQDVLRTRV KTSGIFETKF QVDKVNFHMF DVGAQRDERR KWIQCFNDVT AIIFVVDSSD YNRLQEALND FK SIWNNRW LRTISVILFL NKQDLLAEKV LAGKSKIEDY FPEFARYTTP EDATPEPGED PRVTRAKYFI RDEFLRISTA SGD GRHYCY PHFTCSVDTE NARRIFNDCR DIIQRMHLRQ YELL |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.146707 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWNG SS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Soluble cytochrome b562,D(1B) dopamine receptor
| Macromolecule | Name: Soluble cytochrome b562,D(1B) dopamine receptor / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 68.847297 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: DYKDDDDAKL QTMHHHHHHH HHHADLEDNW ETLNDNLKVI EKADNAAQVK DALTKMRAAA LDAQKATPPK LEDKSPDSPE MKDFRHGFD ILVGQIDDAL KLANEGKVKE AQAAAEQLKT TRNAYIQKYL ASENLYFQGG TMLPPGSNGT AYPGQFALYQ Q LAQGNAVG ...String: DYKDDDDAKL QTMHHHHHHH HHHADLEDNW ETLNDNLKVI EKADNAAQVK DALTKMRAAA LDAQKATPPK LEDKSPDSPE MKDFRHGFD ILVGQIDDAL KLANEGKVKE AQAAAEQLKT TRNAYIQKYL ASENLYFQGG TMLPPGSNGT AYPGQFALYQ Q LAQGNAVG GSAGAPPLGP SQVVTACLLT LLIIWTLLGN VLVCAAIVRS RHLRANMTNV FIVSLAVSDL FVALLVMPWK AV AEVAGYW PFGAFCDVWV AFDIMCSTAS ILNLCVISVD RYWAISRPFR YKRKMTQRMA LVMVGLAWTL SILISFIPVQ LNW HRDQAA SWGGLDLPNN LANWTPWEED FWEPDVNAEN CDSSLNRTYA ISSSLISFYI PVAIMIVTYT RIYRIAQVQI RRIS SLERA AEHAQSCRSS AACAPDTSLR ASIKKETKVL KTLSVIMGVF VCCWLPFFIL NCMVPFCSGH PEGPPAGFPC VSETT FDVF VWFGWANSSL NPVIYAFNAD FQKVFAQLLG CSHFCSRTPV ETVNISNELI SYNQDIVFHK EIAAAYIHMM PNAVTP GNR EVDNDEEEGP FDRMFQIYQT SPDGDPVAES VWELDCEGEI SLDKITPFTP NGFH UniProtKB: Soluble cytochrome b562, D(1B) dopamine receptor |
-Macromolecule #5: Rotigotine
| Macromolecule | Name: Rotigotine / type: ligand / ID: 5 / Number of copies: 1 / Formula: R5F |
|---|---|
| Molecular weight | Theoretical: 315.473 Da |
| Chemical component information | 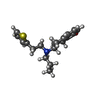 ChemComp-R5F: |
-Macromolecule #6: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 6 / Number of copies: 10 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #7: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 7 / Number of copies: 1 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 4746 / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)