+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3566 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | VipA-N3, non-contractile sheath of the type VI secretion system | |||||||||
 Map data Map data | Postprocessed in Relion2, average resolution is 3.7A. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information: / Type VI secretion system TssC-like / TssC1, N-terminal / TssC1, C-terminal / EvpB/VC_A0108, tail sheath N-terminal domain / EvpB/VC_A0108, tail sheath gpW/gp25-like domain / Type VI secretion system sheath protein TssB1 / Type VI secretion system, VipA, VC_A0107 or Hcp2 / Type VI secretion system effector Hcp / Hcp1-like superfamily / Type VI secretion system effector, Hcp Similarity search - Domain/homology | |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Wang J / Brackmann M / Castano-Diez D / Kudryashev M / Goldie G / Maier T / Stahlberg H / Basler M | |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2017 Journal: Nat Microbiol / Year: 2017Title: Cryo-EM structure of the extended type VI secretion system sheath-tube complex. Authors: Jing Wang / Maximilian Brackmann / Daniel Castaño-Díez / Mikhail Kudryashev / Kenneth N Goldie / Timm Maier / Henning Stahlberg / Marek Basler /   Abstract: The bacterial type VI secretion system (T6SS) uses contraction of a long sheath to quickly thrust a tube with associated effectors across membranes of eukaryotic and bacterial cells . Only limited ...The bacterial type VI secretion system (T6SS) uses contraction of a long sheath to quickly thrust a tube with associated effectors across membranes of eukaryotic and bacterial cells . Only limited structural information is available about the inherently unstable precontraction state of the T6SS. Here, we obtain a 3.7 Å resolution structure of a non-contractile sheath-tube complex using cryo-electron microscopy and show that it resembles the extended T6SS inside Vibrio cholerae cells. We build a pseudo-atomic model of the complete sheath-tube assembly, which provides a mechanistic understanding of coupling sheath contraction with pushing and rotating the inner tube for efficient target membrane penetration. Our data further show that sheath contraction exposes a buried recognition domain to specifically trigger the disassembly and recycling of the T6SS sheath by the cognate ATP-dependent unfoldase ClpV. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3566.map.gz emd_3566.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3566-v30.xml emd-3566-v30.xml emd-3566.xml emd-3566.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3566.png emd_3566.png | 361.8 KB | ||
| Others |  emd_3566_additional.map.gz emd_3566_additional.map.gz | 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3566 http://ftp.pdbj.org/pub/emdb/structures/EMD-3566 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3566 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3566 | HTTPS FTP |
-Related structure data
| Related structure data |  5mxnMC  5ojqMC  3563C  3564C  3567C  5myuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10086 (Title: Cryo EM of Type VI Secretion System VipA-N3/VipB/Hcp complex EMPIAR-10086 (Title: Cryo EM of Type VI Secretion System VipA-N3/VipB/Hcp complexData size: 26.2 Data #1: Unaligned multi-frame micrographs of T6SS VipA-N3/VipB/Hcp complex [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3566.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3566.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed in Relion2, average resolution is 3.7A. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.039 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
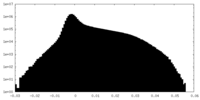
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: low-pass filtered to 5A to visualize the low-resolution...
| File | emd_3566_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | low-pass filtered to 5A to visualize the low-resolution density region of the map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
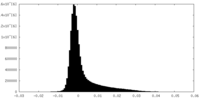
| Density Histograms |
- Sample components
Sample components
-Entire : vipA-N3/vipB noncontractile sheath with Hcp tube
| Entire | Name: vipA-N3/vipB noncontractile sheath with Hcp tube |
|---|---|
| Components |
|
-Supramolecule #1: vipA-N3/vipB noncontractile sheath with Hcp tube
| Supramolecule | Name: vipA-N3/vipB noncontractile sheath with Hcp tube / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Hcp
| Macromolecule | Name: Hcp / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MPTPCYISIE GQTQGLITAG ACTADSIGDS FVEGHEDEML VQQFDHVVTV PTDPQSGQPS GQRVHKPFKF TVALNKAVPL LYNALSSGEK LKTVELKWYR TSIEGKQENF FTTKLENASI VDIHCEMPHC QDPAKSDFTQ NVTVSLSYRK ITWDHVNAGT SGSDDWRKPI EA |
-Macromolecule #2: VipA
| Macromolecule | Name: VipA / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Sequence | String: SKEGSVAPKE RINIKYIPAT GDAQAEVAEV ELPLKTLVVG DFKGHAEQTP LEERATVTVD KNNFEAVMR ESELKITATV KNKLTDDENA ELPVELNFKS LADFAPDAVA SQVPELKKLI E LREALVAL KGPLGNIPAF RERLQSLLNS EESREKLLAE LNL |
-Macromolecule #3: vipB
| Macromolecule | Name: vipB / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MMSTTEKVLE RPQLAQGSLL DEIMAQTRIA PSEEGYDIAK KGVAAFIENL MGSQHSAEPV NKSLVDQMLV ELDKKISAQ MDEILHNSQF QAMESAWRGL KLFVDRTDFR ENNKVEILHV TKDELLEDFE FAPETAQSGL Y KHVYSAGY GQFGGEPVGA IIGNYAFTPS ...String: MMSTTEKVLE RPQLAQGSLL DEIMAQTRIA PSEEGYDIAK KGVAAFIENL MGSQHSAEPV NKSLVDQMLV ELDKKISAQ MDEILHNSQF QAMESAWRGL KLFVDRTDFR ENNKVEILHV TKDELLEDFE FAPETAQSGL Y KHVYSAGY GQFGGEPVGA IIGNYAFTPS TPDMKLLQYM GALGAMAHAP FISSVGPEFF GIDSFEELPN IK DLKSTFE SPKYTKWRSL RESEDARYLG LTAPRFLLRV PYDPIENPVK SFNYAENVSA SHEHYLWGNT AFA FATRLT DSFAKYRWCP NIIGPQSGGA VEDLPVHVFE SMGALQSKIP TEVLITDRKE FELAEEGFIA LTMR KGSDN AAFFSANSIQ KPKVFPNTKE GKEAETNYKL GTQLPYMMII NRLAHYVKVL QREQIGAWKE RQDLE RELN SWIKQYVADQ ENPPADVRSR RPLRAARIEV MDVEGNPGWY QVSLSVRPHF KYMGANFELS LVGRLD QA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-5mxn:  PDB-5ojq: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)