+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of HCA2-Gi complex with acifran | |||||||||
 Map data Map data | map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationActivation of the phototransduction cascade / Olfactory Signaling Pathway / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Prostacyclin signalling through prostacyclin receptor / G alpha (z) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 ...Activation of the phototransduction cascade / Olfactory Signaling Pathway / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Prostacyclin signalling through prostacyclin receptor / G alpha (z) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / G beta:gamma signalling through BTK / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Thromboxane signalling through TP receptor / Thrombin signalling through proteinase activated receptors (PARs) / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / nicotinic acid receptor activity / Hydroxycarboxylic acid-binding receptors / Ca2+ pathway / G alpha (s) signalling events / G alpha (q) signalling events / Extra-nuclear estrogen signaling / G alpha (12/13) signalling events / neutrophil apoptotic process / Vasopressin regulates renal water homeostasis via Aquaporins / GPER1 signaling / G alpha (i) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / positive regulation of neutrophil apoptotic process / ADP signalling through P2Y purinoceptor 1 / Class A/1 (Rhodopsin-like receptors) / phototransduction, visible light / alkylglycerophosphoethanolamine phosphodiesterase activity / positive regulation of adiponectin secretion / photoreceptor outer segment membrane / spectrin binding / photoreceptor outer segment / T cell migration / D2 dopamine receptor binding / Adenylate cyclase inhibitory pathway / negative regulation of lipid catabolic process / positive regulation of protein localization to cell cortex / regulation of cAMP-mediated signaling / G protein-coupled serotonin receptor binding / cellular response to forskolin / regulation of mitotic spindle organization / cardiac muscle cell apoptotic process / photoreceptor inner segment / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / Regulation of insulin secretion / G protein-coupled receptor binding / electron transport chain / G-protein beta/gamma-subunit complex binding / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / response to peptide hormone / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / G alpha (z) signalling events / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / GPER1 signaling / GDP binding / G-protein beta-subunit binding / heterotrimeric G-protein complex / cell junction / signaling receptor complex adaptor activity / GTPase binding / myelin sheath / retina development in camera-type eye / cell cortex / phospholipase C-activating G protein-coupled receptor signaling pathway / positive regulation of cytosolic calcium ion concentration / cellular response to hypoxia / midbody / cell body / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / cell population proliferation / Extra-nuclear estrogen signaling / periplasmic space / electron transfer activity / iron ion binding / G protein-coupled receptor signaling pathway / lysosomal membrane / cell division / GTPase activity / centrosome / dendrite / heme binding / synapse / protein-containing complex binding / GTP binding / nucleolus / magnesium ion binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) / Homo sapiens (human) / synthetic construct (others) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.11 Å | |||||||||
 Authors Authors | Suzuki S / Nishikawa K / Suzuki H / Fujiyoshi Y | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding. Authors: Shota Suzuki / Kotaro Tanaka / Kouki Nishikawa / Hiroshi Suzuki / Atsunori Oshima / Yoshinori Fujiyoshi /  Abstract: Hydroxycarboxylic acid receptors (HCA) are expressed in various tissues and immune cells. HCA2 and its agonist are thus important targets for treating inflammatory and metabolic disorders. Only ...Hydroxycarboxylic acid receptors (HCA) are expressed in various tissues and immune cells. HCA2 and its agonist are thus important targets for treating inflammatory and metabolic disorders. Only limited information is available, however, on the active-state binding of HCAs with agonists. Here, we present cryo-EM structures of human HCA2-Gi and HCA3-Gi signaling complexes binding with multiple compounds bound. Agonists were revealed to form a salt bridge with arginine, which is conserved in the HCA family, to activate these receptors. Extracellular regions of the receptors form a lid-like structure that covers the ligand-binding pocket. Although transmembrane (TM) 6 in HCAs undergoes dynamic conformational changes, ligands do not directly interact with amino acids in TM6, suggesting that indirect signaling induces a slight shift in TM6 to activate Gi proteins. Structural analyses of agonist-bound HCA2 and HCA3 together with mutagenesis and molecular dynamics simulation provide molecular insights into HCA ligand recognition and activation mechanisms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35445.map.gz emd_35445.map.gz | 19.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35445-v30.xml emd-35445-v30.xml emd-35445.xml emd-35445.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35445_fsc.xml emd_35445_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_35445.png emd_35445.png | 42.1 KB | ||
| Masks |  emd_35445_msk_1.map emd_35445_msk_1.map | 20.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-35445.cif.gz emd-35445.cif.gz | 7.1 KB | ||
| Others |  emd_35445_half_map_1.map.gz emd_35445_half_map_1.map.gz emd_35445_half_map_2.map.gz emd_35445_half_map_2.map.gz | 19.3 MB 19.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35445 http://ftp.pdbj.org/pub/emdb/structures/EMD-35445 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35445 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35445 | HTTPS FTP |
-Validation report
| Summary document |  emd_35445_validation.pdf.gz emd_35445_validation.pdf.gz | 941.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35445_full_validation.pdf.gz emd_35445_full_validation.pdf.gz | 940.6 KB | Display | |
| Data in XML |  emd_35445_validation.xml.gz emd_35445_validation.xml.gz | 14.9 KB | Display | |
| Data in CIF |  emd_35445_validation.cif.gz emd_35445_validation.cif.gz | 19.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35445 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35445 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35445 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35445 | HTTPS FTP |
-Related structure data
| Related structure data |  8ihiMC  8ihbC  8ihfC  8ihhC  8ihjC  8ihkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35445.map.gz / Format: CCP4 / Size: 20.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35445.map.gz / Format: CCP4 / Size: 20.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.99 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_35445_msk_1.map emd_35445_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_35445_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_35445_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Multiprotein complex
| Entire | Name: Multiprotein complex |
|---|---|
| Components |
|
-Supramolecule #1: Multiprotein complex
| Supramolecule | Name: Multiprotein complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #5, #1, #3-#4, #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.446047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV AAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY QEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #2: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 26.466486 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL K |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.772562 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHE NLYFQGSSEL DQLRQEAEQL KNQIRDARKA CADATLSQIT NNIDPVGRIQ MRTRRTLRGH LAKIYAMHWG TDSRLLVSA SQDGKLIIWD SYTTNKVHAI PLRSSWVMTC AYAPSGNYVA CGGLDNICSI YNLKTREGNV RVSRELAGHT G YLSCCRFL ...String: MHHHHHHHHE NLYFQGSSEL DQLRQEAEQL KNQIRDARKA CADATLSQIT NNIDPVGRIQ MRTRRTLRGH LAKIYAMHWG TDSRLLVSA SQDGKLIIWD SYTTNKVHAI PLRSSWVMTC AYAPSGNYVA CGGLDNICSI YNLKTREGNV RVSRELAGHT G YLSCCRFL DDNQIVTSSG DTTCALWDIE TGQQTTTFTG HTGDVMSLSL APDTRLFVSG ACDASAKLWD VREGMCRQTF TG HESDINA ICFFPNGNAF ATGSDDATCR LFDLRADQEL MTYSHDNIIC GITSVSFSKS GRLLLAGYDD FNCNVWDALK ADR AGVLAG HDNRVSCLGV TDDGMAVATG SWDSFLKIWN GGSGGGGSGG SSSGGVSGWR LFKKIS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.729947 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ASNNTASIAQ ARKLVEQLKM EANIDRIKVS KAAADLMAYC EAHAKEDPLL TPVPASENPF REKKFFCAIL UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: Soluble cytochrome b562,Hydroxycarboxylic acid receptor 2
| Macromolecule | Name: Soluble cytochrome b562,Hydroxycarboxylic acid receptor 2 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 74.879688 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKADLEDN WETLNDNLKV IEKADNAAQV KDALTKMRAA ALDAQKATPP KLEDKSPDSP EMKDFRHGF DILVGQIDDA LKLANEGKVK EAQAAAEQLK TTRNAYIQKY LNRHHLQDHF LEIDKKNCCV FRDDFIVKVL P PVLGLEFI ...String: MKTIIALSYI FCLVFADYKD DDDKADLEDN WETLNDNLKV IEKADNAAQV KDALTKMRAA ALDAQKATPP KLEDKSPDSP EMKDFRHGF DILVGQIDDA LKLANEGKVK EAQAAAEQLK TTRNAYIQKY LNRHHLQDHF LEIDKKNCCV FRDDFIVKVL P PVLGLEFI FGLLGNGLAL WIFCFHLKSW KSSRIFLFNL AVADFLLIIC LPFLMDNYVR RWDWKFGDIP CRLMLFMLAM NR QGSIIFL TVVAVDRYFR VVHPHHALNK ISNRTAAIIS CLLWGITIGL TVHLLKKKMP IQNGGANLCS SFSICHTFQW HEA MFLLEF FLPLGIILFC SARIIWSLRQ RQMDRHAKIK RAITFIMVVA IVFVICFLPS VVVRIRIFWL LHTSGTQNCE VYRS VDLAF FITLSFTYMN SMLDPVVYYF SSPSFPNFFS TLINRCLQRK MTGEPDNNRS TSVELTGDPN KTRGAPEALM ANSGE PWSP SYLGPTSPEN LYFQGSVFTL EDFVGDWEQT AAYNLDQVLE QGGVSSLLQN LAVSVTPIQR IVRSGENALK IDIHVI IPY EGLSADQMAQ IEEVFKVVYP VDDHHFKVIL PYGTLVIDGV TPNMLNYFGR PYEGIAVFDG KKITVTGTLW NGNKIID ER LITPDGSMLF RVTINS UniProtKB: Soluble cytochrome b562, Hydroxycarboxylic acid receptor 2 |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #7: (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid
| Macromolecule | Name: (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid type: ligand / ID: 7 / Number of copies: 1 / Formula: P9X |
|---|---|
| Molecular weight | Theoretical: 218.205 Da |
| Chemical component information | 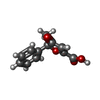 ChemComp-P9X: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 15 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 49.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
 Movie
Movie Controller
Controller


































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)