+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 Delta S-RBD-ACE2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / Delta / RBD / ACE2 / PROTEIN BINDING | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of amino acid transport / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / positive regulation of gap junction assembly / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / regulation of cardiac conduction / maternal process involved in female pregnancy ...positive regulation of amino acid transport / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / positive regulation of gap junction assembly / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / regulation of cardiac conduction / maternal process involved in female pregnancy / regulation of vasoconstriction / peptidyl-dipeptidase activity / transporter activator activity / Metabolism of Angiotensinogen to Angiotensins / carboxypeptidase activity / angiotensin maturation / Attachment and Entry / receptor-mediated endocytosis of virus by host cell / metallocarboxypeptidase activity / viral life cycle / positive regulation of cardiac muscle contraction / regulation of cytokine production / blood vessel diameter maintenance / negative regulation of smooth muscle cell proliferation / brush border membrane / negative regulation of ERK1 and ERK2 cascade / positive regulation of reactive oxygen species metabolic process / metallopeptidase activity / endocytic vesicle membrane / regulation of cell population proliferation / virus receptor activity / regulation of inflammatory response / endopeptidase activity / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Potential therapeutics for SARS / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / membrane fusion / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / cilium / symbiont-mediated suppression of host innate immune response / apical plasma membrane / membrane raft / receptor ligand activity / endocytosis involved in viral entry into host cell / endoplasmic reticulum lumen / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / cell surface / negative regulation of transcription by RNA polymerase II / extracellular space / extracellular exosome / extracellular region / zinc ion binding / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Xu J / Meng F / Liu N / Wang HW | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Self-assembled superstructure alleviates air-water interface effect in cryo-EM. Authors: Liming Zheng / Jie Xu / Weihua Wang / Xiaoyin Gao / Chao Zhao / Weijun Guo / Luzhao Sun / Hang Cheng / Fanhao Meng / Buhang Chen / Weiyu Sun / Xia Jia / Xiong Zhou / Kai Wu / Zhongfan Liu / ...Authors: Liming Zheng / Jie Xu / Weihua Wang / Xiaoyin Gao / Chao Zhao / Weijun Guo / Luzhao Sun / Hang Cheng / Fanhao Meng / Buhang Chen / Weiyu Sun / Xia Jia / Xiong Zhou / Kai Wu / Zhongfan Liu / Feng Ding / Nan Liu / Hong-Wei Wang / Hailin Peng /  Abstract: Cryo-electron microscopy (cryo-EM) has been widely used to reveal the structures of proteins at atomic resolution. One key challenge is that almost all proteins are predominantly adsorbed to the air- ...Cryo-electron microscopy (cryo-EM) has been widely used to reveal the structures of proteins at atomic resolution. One key challenge is that almost all proteins are predominantly adsorbed to the air-water interface during standard cryo-EM specimen preparation. The interaction of proteins with air-water interface will significantly impede the success of reconstruction and achievable resolution. Here, we highlight the critical role of impenetrable surfactant monolayers in passivating the air-water interface problems, and develop a robust effective method for high-resolution cryo-EM analysis, by using the superstructure GSAMs which comprises surfactant self-assembled monolayers (SAMs) and graphene membrane. The GSAMs works well in enriching the orientations and improving particle utilization ratio of multiple proteins, facilitating the 3.3-Å resolution reconstruction of a 100-kDa protein complex (ACE2-RBD), which shows strong preferential orientation using traditional specimen preparation protocol. Additionally, we demonstrate that GSAMs enables the successful determinations of small proteins (<100 kDa) at near-atomic resolution. This study expands the understanding of SAMs and provides a key to better control the interaction of protein with air-water interface. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34977.map.gz emd_34977.map.gz | 117.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34977-v30.xml emd-34977-v30.xml emd-34977.xml emd-34977.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34977.png emd_34977.png | 72 KB | ||
| Filedesc metadata |  emd-34977.cif.gz emd-34977.cif.gz | 5.9 KB | ||
| Others |  emd_34977_half_map_1.map.gz emd_34977_half_map_1.map.gz emd_34977_half_map_2.map.gz emd_34977_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34977 http://ftp.pdbj.org/pub/emdb/structures/EMD-34977 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34977 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34977 | HTTPS FTP |
-Validation report
| Summary document |  emd_34977_validation.pdf.gz emd_34977_validation.pdf.gz | 804.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34977_full_validation.pdf.gz emd_34977_full_validation.pdf.gz | 804.1 KB | Display | |
| Data in XML |  emd_34977_validation.xml.gz emd_34977_validation.xml.gz | 14 KB | Display | |
| Data in CIF |  emd_34977_validation.cif.gz emd_34977_validation.cif.gz | 16.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34977 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34977 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34977 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34977 | HTTPS FTP |
-Related structure data
| Related structure data |  8hrlMC  8hriC  8hrjC  8hrkC  8hrmC  8hrnC  8hruC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34977.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34977.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
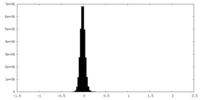
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34977_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
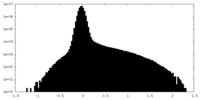
| Density Histograms |
-Half map: #1
| File | emd_34977_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
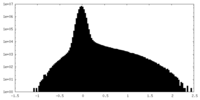
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 Delta S-RBD-ACE2 complex
| Entire | Name: SARS-CoV-2 Delta S-RBD-ACE2 complex |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Delta S-RBD-ACE2 complex
| Supramolecule | Name: SARS-CoV-2 Delta S-RBD-ACE2 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: on graphene membranes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Processed angiotensin-converting enzyme 2
| Macromolecule | Name: Processed angiotensin-converting enzyme 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 69.153664 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: STIEEQAKTF LDKFNHEAED LFYQSSLASW NYNTNITEEN VQNMNNAGDK WSAFLKEQST LAQMYPLQEI QNLTVKLQLQ ALQQNGSSV LSEDKSKRLN TILNTMSTIY STGKVCNPDN PQECLLLEPG LNEIMANSLD YNERLWAWES WRSEVGKQLR P LYEEYVVL ...String: STIEEQAKTF LDKFNHEAED LFYQSSLASW NYNTNITEEN VQNMNNAGDK WSAFLKEQST LAQMYPLQEI QNLTVKLQLQ ALQQNGSSV LSEDKSKRLN TILNTMSTIY STGKVCNPDN PQECLLLEPG LNEIMANSLD YNERLWAWES WRSEVGKQLR P LYEEYVVL KNEMARANHY EDYGDYWRGD YEVNGVDGYD YSRGQLIEDV EHTFEEIKPL YEHLHAYVRA KLMNAYPSYI SP IGCLPAH LLGDMWGRFW TNLYSLTVPF GQKPNIDVTD AMVDQAWDAQ RIFKEAEKFF VSVGLPNMTQ GFWENSMLTD PGN VQKAVC HPTAWDLGKG DFRILMCTKV TMDDFLTAHH EMGHIQYDMA YAAQPFLLRN GANEGFHEAV GEIMSLSAAT PKHL KSIGL LSPDFQEDNE TEINFLLKQA LTIVGTLPFT YMLEKWRWMV FKGEIPKDQW MKKWWEMKRE IVGVVEPVPH DETYC DPAS LFHVSNDYSF IRYYTRTLYQ FQFQEALCQA AKHEGPLHKC DISNSTEAGQ KLFNMLRLGK SEPWTLALEN VVGAKN MNV RPLLNYFEPL FTWLKDQNKN SFVGWSTDWS PYAD UniProtKB: Angiotensin-converting enzyme 2 |
-Macromolecule #2: Spike protein S1
| Macromolecule | Name: Spike protein S1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.848492 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TNLCPFGEVF NATRFASVYA WNRKRISNCV ADYSVLYNSA SFSTFKCYGV SPTKLNDLCF TNVYADSFVI RGDEVRQIAP GQTGKIADY NYKLPDDFTG CVIAWNSNNL DSKVGGNYNY RYRLFRKSNL KPFERDISTE IYQAGSKPCN GVEGFNCYFP L QSYGFQPT ...String: TNLCPFGEVF NATRFASVYA WNRKRISNCV ADYSVLYNSA SFSTFKCYGV SPTKLNDLCF TNVYADSFVI RGDEVRQIAP GQTGKIADY NYKLPDDFTG CVIAWNSNNL DSKVGGNYNY RYRLFRKSNL KPFERDISTE IYQAGSKPCN GVEGFNCYFP L QSYGFQPT NGVGYQPYRV VVLSFELLHA PATVCG UniProtKB: Spike glycoprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 224616 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)