+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of streptavidin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Streptomyces avidinii (bacteria) Streptomyces avidinii (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.56 Å | |||||||||
 Authors Authors | Xu J / Liu N / Wang HW | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Self-assembled superstructure alleviates air-water interface effect in cryo-EM. Authors: Liming Zheng / Jie Xu / Weihua Wang / Xiaoyin Gao / Chao Zhao / Weijun Guo / Luzhao Sun / Hang Cheng / Fanhao Meng / Buhang Chen / Weiyu Sun / Xia Jia / Xiong Zhou / Kai Wu / Zhongfan Liu / ...Authors: Liming Zheng / Jie Xu / Weihua Wang / Xiaoyin Gao / Chao Zhao / Weijun Guo / Luzhao Sun / Hang Cheng / Fanhao Meng / Buhang Chen / Weiyu Sun / Xia Jia / Xiong Zhou / Kai Wu / Zhongfan Liu / Feng Ding / Nan Liu / Hong-Wei Wang / Hailin Peng /  Abstract: Cryo-electron microscopy (cryo-EM) has been widely used to reveal the structures of proteins at atomic resolution. One key challenge is that almost all proteins are predominantly adsorbed to the air- ...Cryo-electron microscopy (cryo-EM) has been widely used to reveal the structures of proteins at atomic resolution. One key challenge is that almost all proteins are predominantly adsorbed to the air-water interface during standard cryo-EM specimen preparation. The interaction of proteins with air-water interface will significantly impede the success of reconstruction and achievable resolution. Here, we highlight the critical role of impenetrable surfactant monolayers in passivating the air-water interface problems, and develop a robust effective method for high-resolution cryo-EM analysis, by using the superstructure GSAMs which comprises surfactant self-assembled monolayers (SAMs) and graphene membrane. The GSAMs works well in enriching the orientations and improving particle utilization ratio of multiple proteins, facilitating the 3.3-Å resolution reconstruction of a 100-kDa protein complex (ACE2-RBD), which shows strong preferential orientation using traditional specimen preparation protocol. Additionally, we demonstrate that GSAMs enables the successful determinations of small proteins (<100 kDa) at near-atomic resolution. This study expands the understanding of SAMs and provides a key to better control the interaction of protein with air-water interface. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34978.map.gz emd_34978.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34978-v30.xml emd-34978-v30.xml emd-34978.xml emd-34978.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34978.png emd_34978.png | 87.4 KB | ||
| Filedesc metadata |  emd-34978.cif.gz emd-34978.cif.gz | 5.1 KB | ||
| Others |  emd_34978_half_map_1.map.gz emd_34978_half_map_1.map.gz emd_34978_half_map_2.map.gz emd_34978_half_map_2.map.gz | 27.2 MB 27.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34978 http://ftp.pdbj.org/pub/emdb/structures/EMD-34978 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34978 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34978 | HTTPS FTP |
-Validation report
| Summary document |  emd_34978_validation.pdf.gz emd_34978_validation.pdf.gz | 832.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34978_full_validation.pdf.gz emd_34978_full_validation.pdf.gz | 832.2 KB | Display | |
| Data in XML |  emd_34978_validation.xml.gz emd_34978_validation.xml.gz | 10.8 KB | Display | |
| Data in CIF |  emd_34978_validation.cif.gz emd_34978_validation.cif.gz | 12.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34978 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34978 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34978 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34978 | HTTPS FTP |
-Related structure data
| Related structure data |  8hrmMC  8hriC  8hrjC  8hrkC  8hrlC  8hrnC  8hruC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34978.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34978.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.5191 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34978_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
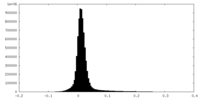
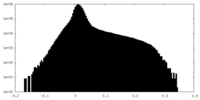
| Density Histograms |
-Half map: #2
| File | emd_34978_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
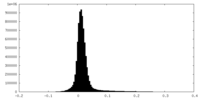
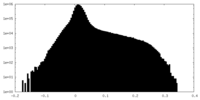
| Density Histograms |
- Sample components
Sample components
-Entire : streptavidin
| Entire | Name: streptavidin |
|---|---|
| Components |
|
-Supramolecule #1: streptavidin
| Supramolecule | Name: streptavidin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: on graphene/SAMs membranes |
|---|---|
| Source (natural) | Organism:  Streptomyces avidinii (bacteria) Streptomyces avidinii (bacteria) |
-Macromolecule #1: Streptavidin
| Macromolecule | Name: Streptavidin / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptomyces avidinii (bacteria) Streptomyces avidinii (bacteria) |
| Molecular weight | Theoretical: 12.596641 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GITGTWYNQL GSTFIVTAGA DGALTGTYES AVGNAESRYV LTGRYDSAPA TDGSGTALGW TVAWKNNYRN AHSATTWSGQ YVGGAEARI NTQWLLTSGT TEANAWKSTL VGHDTFTKVK UniProtKB: Streptavidin |
-Macromolecule #2: water
| Macromolecule | Name: water / type: ligand / ID: 2 / Number of copies: 19 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.56 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 99871 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)