+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3458 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | negative-stain volume of Sso DNA PolB1 | |||||||||
 Map data Map data | Final map from iteration 10 onto which FSC has been calculated and displayed (Figure S4C, right). Contour level in Chimera | |||||||||
 Sample Sample |
| |||||||||
| Biological species | Archaea (unknown) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 22.8 Å | |||||||||
 Authors Authors | Abrescia NGA / Bell SD | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: Identification and characterization of a heterotrimeric archaeal DNA polymerase holoenzyme. Authors: Jiangyu Yan / Thomas R Beattie / Adriana L Rojas / Kelly Schermerhorn / Tamzin Gristwood / Jonathan C Trinidad / Sonja V Albers / Pietro Roversi / Andrew F Gardner / Nicola G A Abrescia / Stephen D Bell /     Abstract: Since their initial characterization over 30 years ago, it has been believed that the archaeal B-family DNA polymerases are single-subunit enzymes. This contrasts with the multi-subunit B-family ...Since their initial characterization over 30 years ago, it has been believed that the archaeal B-family DNA polymerases are single-subunit enzymes. This contrasts with the multi-subunit B-family replicative polymerases of eukaryotes. Here we reveal that the highly studied PolB1 from Sulfolobus solfataricus exists as a heterotrimeric complex in cell extracts. Two small subunits, PBP1 and PBP2, associate with distinct surfaces of the larger catalytic subunit and influence the enzymatic properties of the DNA polymerase. Thus, multi-subunit replicative DNA polymerase holoenzymes are present in all three domains of life. We reveal the architecture of the assembly by a combination of cross-linking coupled with mass spectrometry, X-ray crystallography and single-particle electron microscopy. The small subunits stabilize the holoenzyme assembly and the acidic tail of one small subunit mitigates the ability of the enzyme to perform strand-displacement synthesis, with important implications for lagging strand DNA synthesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3458.map.gz emd_3458.map.gz | 6.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3458-v30.xml emd-3458-v30.xml emd-3458.xml emd-3458.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3458.png emd_3458.png | 111.1 KB | ||
| Masks |  emd_3458_msk_1.map emd_3458_msk_1.map | 8 MB |  Mask map Mask map | |
| Others |  emd_3458_additional.map.gz emd_3458_additional.map.gz emd_3458_half_map_1.map.gz emd_3458_half_map_1.map.gz emd_3458_half_map_2.map.gz emd_3458_half_map_2.map.gz | 874.8 KB 6.1 MB 6.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3458 http://ftp.pdbj.org/pub/emdb/structures/EMD-3458 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3458 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3458 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3458.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3458.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final map from iteration 10 onto which FSC has been calculated and displayed (Figure S4C, right). Contour level in Chimera | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.66 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
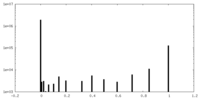
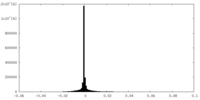
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_3458_msk_1.map emd_3458_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
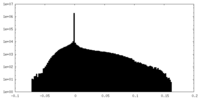
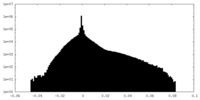
| Density Histograms |
-Additional map: This is the masked map used in Figure...
| File | emd_3458_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the masked map used in Figure 4A and in submitted figure. Contour level in Chimera. | ||||||||||||
| Projections & Slices |
| ||||||||||||
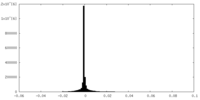
| Density Histograms |
-Half map: half2 map from iteration 10. Contour level in Chimera
| File | emd_3458_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 map from iteration 10. Contour level in Chimera | ||||||||||||
| Projections & Slices |
| ||||||||||||
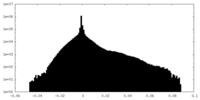
| Density Histograms |
-Half map: half1 map from iteration 10. Contour level in Chimera
| File | emd_3458_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 map from iteration 10. Contour level in Chimera | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : archaeal DNA polymerase B1
| Entire | Name: archaeal DNA polymerase B1 |
|---|---|
| Components |
|
-Supramolecule #1: archaeal DNA polymerase B1
| Supramolecule | Name: archaeal DNA polymerase B1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: This is the apo enzyme. |
|---|---|
| Source (natural) | Organism: Archaea (unknown) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 101 KDa |
-Macromolecule #1: DNA polymerase B1
| Macromolecule | Name: DNA polymerase B1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: Archaea (unknown) |
| Recombinant expression | Organism:  |
| Sequence | String: MAKQLTLFDI PSSKPAKSEQ NTQQSQQSAP VEEKKVVRRE WLEEAQENKI YFLLQVDYDG KKGKAVCKLF DKETQKIYAL YDNTGHKPYF LVDLEPDKVG KIPKIVRDPS FDHIETVSKI DPYTWNKFKL TKIVVRDPLA VRRLRNDVPK AYEAHIKYFN NYMYDIGLIP ...String: MAKQLTLFDI PSSKPAKSEQ NTQQSQQSAP VEEKKVVRRE WLEEAQENKI YFLLQVDYDG KKGKAVCKLF DKETQKIYAL YDNTGHKPYF LVDLEPDKVG KIPKIVRDPS FDHIETVSKI DPYTWNKFKL TKIVVRDPLA VRRLRNDVPK AYEAHIKYFN NYMYDIGLIP GMPYVVKNGK LESVYLSLDE KDVEEIKKAF ADSDEMTRQM AVDWLPIFET EIPKIKRVAI DIEVYTPVKG RIPDSQKAEF PIISIALAGS DGLKKVLVLN RNDVNEGSVK LDGISVERFN TEYELLGRFF DILLEYPIVL TFNGDDFDLP YIYFRALKLG YFPEEIPIDV AGKDEAKYLA GLHIDLYKFF FNKAVRNYAF EGKYNEYNLD AVAKALLGTS KVKVDTLISF LDVEKLIEYN FRDAEITLQL TTFNNDLTMK LIVLFSRISR LGIEELTRTE ISTWVKNLYY WEHRKRNWLI PLKEEILAKS SNIRTSALIK GKGYKGAVVI DPPAGIFFNI TVLDFASLYP SIIRTWNLSY ETVDIQQCKK PYEVKDETGE VLHIVCMDRP GITAVITGLL RDFRVKIYKK KAKNPNNSEE QKLLYDVVQR AMKVFINATY GVFGAETFPL YAPAVAESVT ALGRYVITST VKKAREEGLT VLYGDTDSLF LLNPPKNSLE NIIKWVKTTF NLDLEVDKTY KFVAFSGLKK NYFGVYQDGK VDIKGMLVKK RNTPEFVKKV FNEVKELMIS INSPNDVKEI KRKIVDVVKG SYEKLKNKGY NLDELAFKVM LSKPLDAYKK NTPQHVKAAL QLRPFGVNVL PRDIIYYVKV RSKDGVKPVQ LAKVTEIDAE KYLEALRSTF EQILRAFGVS WDEIAATMSI DSFFSYPSKG NS |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Staining | Type: NEGATIVE / Material: uranyl formate Details: Negatively stained EM specimens were prepared using carbon-coated grids and stained with 2% of uranyl formate solution. | ||||||||||||
| Grid | Model: EMS / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.02 kPa |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FSC |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 108 / Average exposure time: 0.5 sec. / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 90201 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 1.7 µm / Nominal defocus min: 1.3 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation coefficient |
|---|
 Movie
Movie Controller
Controller



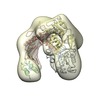









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)