+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3399 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the core NuRD complex (MTA1:HDAC1:RBBP4) | |||||||||
 Map data Map data | Structure of the core NuRD complex: MTA1:HDAC1:RBBP4 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Transcription / HDAC1 / MTA1 / RBBP4 / Chromatin / Histone Deacetylase / Metastasis associated protein / Histone binding protein | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
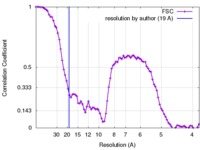
| Method | single particle reconstruction / negative staining / Resolution: 19.0 Å | |||||||||
 Authors Authors | Millard CJ / Saleh A / Morris K / Fairall L / Smith CJ / Schwabe JWR | |||||||||
 Citation Citation |  Journal: Elife / Year: 2016 Journal: Elife / Year: 2016Title: The structure of the core NuRD repression complex provides insights into its interaction with chromatin. Authors: Christopher J Millard / Niranjan Varma / Almutasem Saleh / Kyle Morris / Peter J Watson / Andrew R Bottrill / Louise Fairall / Corinne J Smith / John W R Schwabe /  Abstract: The NuRD complex is a multi-protein transcriptional corepressor that couples histone deacetylase and ATP-dependent chromatin remodelling activities. The complex regulates the higher-order structure ...The NuRD complex is a multi-protein transcriptional corepressor that couples histone deacetylase and ATP-dependent chromatin remodelling activities. The complex regulates the higher-order structure of chromatin, and has important roles in the regulation of gene expression, DNA damage repair and cell differentiation. HDACs 1 and 2 are recruited by the MTA1 corepressor to form the catalytic core of the complex. The histone chaperone protein RBBP4, has previously been shown to bind to the carboxy-terminal tail of MTA1. We show that MTA1 recruits a second copy of RBBP4. The crystal structure reveals an extensive interface between MTA1 and RBBP4. An EM structure, supported by SAXS and crosslinking, reveals the architecture of the dimeric HDAC1:MTA1:RBBP4 assembly which forms the core of the NuRD complex. We find evidence that in this complex RBBP4 mediates interaction with histone H3 tails, but not histone H4, suggesting a mechanism for recruitment of the NuRD complex to chromatin. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3399.map.gz emd_3399.map.gz | 3.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3399-v30.xml emd-3399-v30.xml emd-3399.xml emd-3399.xml | 10.9 KB 10.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3399_fsc.xml emd_3399_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_3399.tif emd_3399.tif | 122.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3399 http://ftp.pdbj.org/pub/emdb/structures/EMD-3399 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3399 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3399 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3399.map.gz / Format: CCP4 / Size: 41.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3399.map.gz / Format: CCP4 / Size: 41.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the core NuRD complex: MTA1:HDAC1:RBBP4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.921 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Core NuRD complex (MTA1:HDAC1:RBBP4)
| Entire | Name: Core NuRD complex (MTA1:HDAC1:RBBP4) |
|---|---|
| Components |
|
-Supramolecule #1000: Core NuRD complex (MTA1:HDAC1:RBBP4)
| Supramolecule | Name: Core NuRD complex (MTA1:HDAC1:RBBP4) / type: sample / ID: 1000 / Details: Dimer / Oligomeric state: 2 / Number unique components: 3 |
|---|---|
| Molecular weight | Experimental: 300 KDa / Theoretical: 300 KDa / Method: SEC-MALS |
-Macromolecule #1: MTA1
| Macromolecule | Name: MTA1 / type: ligand / ID: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism: HEK293F / Recombinant plasmid: pcDNA3 |
-Macromolecule #2: HDAC1
| Macromolecule | Name: HDAC1 / type: ligand / ID: 2 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism: HEK293F / Recombinant plasmid: pcDNA3 |
-Macromolecule #3: RBBP4
| Macromolecule | Name: RBBP4 / type: ligand / ID: 3 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism: HEK293F / Recombinant plasmid: pcDNA3 |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20 mM Tris/HCl (pH 7.5), 40 mM NaCl |
| Staining | Type: NEGATIVE / Details: 2% uranyl acetate for 1 min |
| Grid | Details: Prepared by glow discharging carbon coated copper 300 mesh grids (agar scientific) at 10 mA for 30 seconds |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2010F |
|---|---|
| Date | Jul 22, 2015 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 308 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Fit in map (Chimera) |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Fit in map (Chimera) |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)