+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the galanin-bound GALR1-miniGo complex | |||||||||
 Map data Map data | cryoEM map of the GALR1/Go/galanin complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / Galanin receptor 1 / miniGo / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationgalanin receptor binding / type 1 galanin receptor binding / type 2 galanin receptor binding / type 3 galanin receptor binding / positive regulation of large conductance calcium-activated potassium channel activity / parental behavior / galanin receptor activity / positive regulation of timing of catagen / positive regulation of cortisol secretion / regulation of glucocorticoid metabolic process ...galanin receptor binding / type 1 galanin receptor binding / type 2 galanin receptor binding / type 3 galanin receptor binding / positive regulation of large conductance calcium-activated potassium channel activity / parental behavior / galanin receptor activity / positive regulation of timing of catagen / positive regulation of cortisol secretion / regulation of glucocorticoid metabolic process / negative regulation of lymphocyte proliferation / : / negative regulation of adenylate cyclase activity / neuropeptide hormone activity / mu-type opioid receptor binding / corticotropin-releasing hormone receptor 1 binding / feeding behavior / neuropeptide binding / vesicle docking involved in exocytosis / G protein-coupled dopamine receptor signaling pathway / insulin secretion / regulation of heart contraction / response to immobilization stress / parallel fiber to Purkinje cell synapse / peptide hormone binding / neuropeptide signaling pathway / postsynaptic modulation of chemical synaptic transmission / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / muscle contraction / secretory granule / Peptide ligand-binding receptors / locomotory behavior / negative regulation of insulin secretion / response to insulin / GABA-ergic synapse / response to estrogen / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / nervous system development / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / positive regulation of cytosolic calcium ion concentration / cell body / G protein activity / presynaptic membrane / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / postsynaptic membrane / Extra-nuclear estrogen signaling / cell population proliferation / positive regulation of apoptotic process / G protein-coupled receptor signaling pathway Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Jiang W / Zheng S | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Structural insights into galanin receptor signaling. Authors: Wentong Jiang / Sanduo Zheng /  Abstract: Galanin is a biologically active neuropeptide, and functions through three distinct G protein–coupled receptors (GPCRs), namely GALR1, GALR2, and GALR3. GALR signaling plays important roles in ...Galanin is a biologically active neuropeptide, and functions through three distinct G protein–coupled receptors (GPCRs), namely GALR1, GALR2, and GALR3. GALR signaling plays important roles in regulating various physiological processes such as energy metabolism, neuropathic pain, epileptic activity, and sleep homeostasis. GALR1 and GALR3 signal through the Gi/o pathway, whereas GALR2 signals mainly through the Gq/11 pathway. However, the molecular basis for galanin recognition and G protein selectivity of GALRs remains poorly understood. Here, we report the cryoelectron microscopy structures of the GALR1-Go and the GALR2-Gq complexes bound to the endogenous ligand galanin or spexin. The galanin peptide mainly adopts an alpha helical structure, which binds at the extracellular vestibule of the receptors, nearly parallel to the membrane plane without penetrating deeply into the receptor core. Structural analysis combined with functional studies reveals important structural determinants for the G protein selectivity of GALRs as well as other class A GPCRs. In addition, we show that the zinc ion is a negative allosteric regulator of GALR1 but not GALR2. Our studies provide insight into the mechanisms of G protein selectivity of GPCRs and highlight a potential function of the neuromodulator zinc ion as a modulator of GPCR signaling in the central nervous system. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33229.map.gz emd_33229.map.gz | 20.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33229-v30.xml emd-33229-v30.xml emd-33229.xml emd-33229.xml | 24.3 KB 24.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33229.png emd_33229.png | 29.3 KB | ||
| Filedesc metadata |  emd-33229.cif.gz emd-33229.cif.gz | 7.4 KB | ||
| Others |  emd_33229_half_map_1.map.gz emd_33229_half_map_1.map.gz emd_33229_half_map_2.map.gz emd_33229_half_map_2.map.gz | 20.6 MB 20.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33229 http://ftp.pdbj.org/pub/emdb/structures/EMD-33229 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33229 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33229 | HTTPS FTP |
-Related structure data
| Related structure data |  7xjjMC  7xjkC  7xjlC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33229.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33229.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoEM map of the GALR1/Go/galanin complex | ||||||||||||||||||||||||||||||||||||
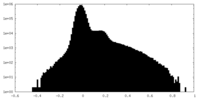
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.087 Å | ||||||||||||||||||||||||||||||||||||
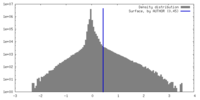
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map of the GALR1/Go/galanin complex
| File | emd_33229_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map of the GALR1/Go/galanin complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
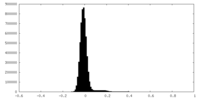
| Density Histograms |
-Half map: half map of the GALR1/Go/galanin complex
| File | emd_33229_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map of the GALR1/Go/galanin complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
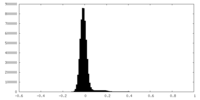
| Density Histograms |
- Sample components
Sample components
-Entire : Galanin-bound Galanin receptor type 1 in complex with miniGalphao...
| Entire | Name: Galanin-bound Galanin receptor type 1 in complex with miniGalphao, Gbeta/gamma subunit and a single-chain variable fragment (scFv16) |
|---|---|
| Components |
|
-Supramolecule #1: Galanin-bound Galanin receptor type 1 in complex with miniGalphao...
| Supramolecule | Name: Galanin-bound Galanin receptor type 1 in complex with miniGalphao, Gbeta/gamma subunit and a single-chain variable fragment (scFv16) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: G protein subunit alpha o1,Guanine nucleotide-binding protein G(o...
| Macromolecule | Name: G protein subunit alpha o1,Guanine nucleotide-binding protein G(o) subunit alpha type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.447115 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGCTLSAEER AALERSKAIE KNLKEDGISA AKDVKLLLLG ADNSGKSTIV KQMKIIHGGS GGSGGTTGIV ETHFTFKNLH FRLFDVGGQ RSERKKWIHC FEDVTAIIFC VDLSDYNRMH ESLMLFDSIC NNKFFIDTSI ILFLNKKDLF GEKIKKSPLT I CFPEYTGP ...String: MGCTLSAEER AALERSKAIE KNLKEDGISA AKDVKLLLLG ADNSGKSTIV KQMKIIHGGS GGSGGTTGIV ETHFTFKNLH FRLFDVGGQ RSERKKWIHC FEDVTAIIFC VDLSDYNRMH ESLMLFDSIC NNKFFIDTSI ILFLNKKDLF GEKIKKSPLT I CFPEYTGP NTYEDAAAYI QAQFESKNRS PNKEIYCHMT CATDTNNAQV IFDAVTDIII ANNLRGCGLY UniProtKB: G protein subunit alpha o1, Guanine nucleotide-binding protein G(o) subunit alpha, Guanine nucleotide-binding protein G(o) subunit alpha |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.41693 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Galanin
| Macromolecule | Name: Galanin / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.161446 KDa |
| Sequence | String: GWTLNSAGYL LGPHAVGNHR SFSDKNGLTS UniProtKB: Galanin peptides |
-Macromolecule #4: Galanin receptor type 1
| Macromolecule | Name: Galanin receptor type 1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.38293 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DYKDDDDKGS MELAVGNLSE GNASWPEPPA PEPGPLFGIG VENFVTLVVF GLIFALGVLG NSLVITVLAR SKPGKPRSTT NLFILNLSI ADLAYLLFCI PFQATVYALP TWVLGAFICK FIHYFFTVSM LVSIFTLAAM SVDRYVAIVH SRRSSSLRVS R NALLGVGC ...String: DYKDDDDKGS MELAVGNLSE GNASWPEPPA PEPGPLFGIG VENFVTLVVF GLIFALGVLG NSLVITVLAR SKPGKPRSTT NLFILNLSI ADLAYLLFCI PFQATVYALP TWVLGAFICK FIHYFFTVSM LVSIFTLAAM SVDRYVAIVH SRRSSSLRVS R NALLGVGC IWALSIAMAS PVAYHQGLFH PRASNQTFCW EQWPDPRHKK AYVVCTFVFG YLLPLLLICF CYAKVLNHLH KK LKNMSKK SEASKKKTAQ TVLVVVVVFG ISWLPHHIIH LWAEFGVFPL TPASFLFRIT AHCLAYSNSS VNPIIYAFLS ENF RKAYKQ VFKCHIGGGG GGAGALEVLF Q UniProtKB: Galanin receptor type 1 |
-Macromolecule #5: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.845078 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFSAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #6: single Fab chain (svFv16)
| Macromolecule | Name: single Fab chain (svFv16) / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 31.857369 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PDVQLVESGG GLVQPGGSRK LSCSASGFAF SSFGMHWVRQ APEKGLEWVA YISSGSGTIY YADTVKGRFT ISRDDPKNTL FLQMTSLRS EDTAMYYCVR SIYYYGSSPF DFWGQGTTLT VSSGGGGSGG GGSGGGGSDI VMTQATSSVP VTPGESVSIS C RSSKSLLH ...String: PDVQLVESGG GLVQPGGSRK LSCSASGFAF SSFGMHWVRQ APEKGLEWVA YISSGSGTIY YADTVKGRFT ISRDDPKNTL FLQMTSLRS EDTAMYYCVR SIYYYGSSPF DFWGQGTTLT VSSGGGGSGG GGSGGGGSDI VMTQATSSVP VTPGESVSIS C RSSKSLLH SNGNTYLYWF LQRPGQSPQL LIYRMSNLAS GVPDRFSGSG SGTAFTLTIS RLEAEDVGVY YCMQHLEYPL TF GAGTKLE LKAAAGAPLE VLFQGPGAWS HPQFEKGAED QVDPRLIDGK GAAHHHHHHH H |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.1 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281.15 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7xjj: |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)