+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32328 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of GmALMT12/QUAC1 anion channel | ||||||||||||
 Map data Map data | Half map 2 for GmALMT12/QUAC1 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Symmetrical dimer / T-shaped pore / twisted two-layer architecture / Malate-modulation / MEMBRANE PROTEIN | ||||||||||||
| Function / homology | malate transport / Aluminum-activated malate transporter / Aluminium activated malate transporter / plant-type vacuole membrane / monoatomic ion transmembrane transport / Aluminum-activated malate transporter 12 Function and homology information Function and homology information | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||
 Authors Authors | Qin L / Tang LH | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Cryo-EM structure and electrophysiological characterization of ALMT from reveal a previously uncharacterized class of anion channels. Authors: Li Qin / Ling-Hui Tang / Jia-Shu Xu / Xian-Hui Zhang / Yun Zhu / Chun-Rui Zhang / Mei-Hua Wang / Xue-Lei Liu / Fei Li / Fei Sun / Min Su / Yujia Zhai / Yu-Hang Chen /   Abstract: Aluminum-activated malate transporters (ALMTs) form an anion channel family that plays essential roles in diverse functions in plants. ALMT12, also named QUAC1 (quick anion channel 1), regulates ...Aluminum-activated malate transporters (ALMTs) form an anion channel family that plays essential roles in diverse functions in plants. ALMT12, also named QUAC1 (quick anion channel 1), regulates stomatal closure in response to environmental stimuli. However, the molecular basis of ALMT12/QUAC1 activity remains elusive. Here, we describe the cryo-EM structure of ALMT12/QUAC1 from at 3.5-Å resolution. ALMT12/QUAC1 is a symmetrical dimer, forming a single electropositive T-shaped pore across the membrane. The transmembrane and cytoplasmic domains are assembled into a twisted two-layer architecture, with their associated dimeric interfaces nearly perpendicular. ALMT12/QUAC1-mediated currents display rapid kinetics of activation/deactivation and a bell-shaped voltage dependency, reminiscent of the rapid (R)-type anion currents. Our structural and functional analyses reveal a domain-twisting mechanism for malate-mediated activation. Together, our study uncovers the molecular basis for a previously uncharacterized class of anion channels and provides insights into the gating and modulation of the ALMT12/QUAC1 anion channel. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32328.map.gz emd_32328.map.gz | 21 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32328-v30.xml emd-32328-v30.xml emd-32328.xml emd-32328.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32328.png emd_32328.png | 69.9 KB | ||
| Filedesc metadata |  emd-32328.cif.gz emd-32328.cif.gz | 6 KB | ||
| Others |  emd_32328_additional_1.map.gz emd_32328_additional_1.map.gz emd_32328_half_map_1.map.gz emd_32328_half_map_1.map.gz emd_32328_half_map_2.map.gz emd_32328_half_map_2.map.gz | 40.3 MB 39.8 MB 39.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32328 http://ftp.pdbj.org/pub/emdb/structures/EMD-32328 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32328 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32328 | HTTPS FTP |
-Related structure data
| Related structure data |  7w6kMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32328.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32328.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 for GmALMT12/QUAC1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Sharpen map for GmALMT12/QUAC1
| File | emd_32328_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
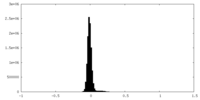
| Annotation | Sharpen map for GmALMT12/QUAC1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
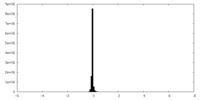
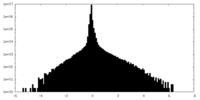
| Density Histograms |
-Half map: Reconstructed Density map for GmALMT12/QUAC1
| File | emd_32328_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
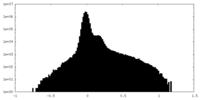
| Annotation | Reconstructed Density map for GmALMT12/QUAC1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
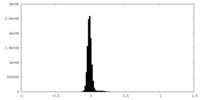
| Density Histograms |
-Half map: Half map 1 for GmALMT12/QUAC1
| File | emd_32328_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 for GmALMT12/QUAC1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
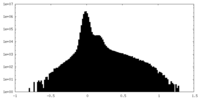
| Density Histograms |
- Sample components
Sample components
-Entire : GmALMT12/QUAC1
| Entire | Name: GmALMT12/QUAC1 |
|---|---|
| Components |
|
-Supramolecule #1: GmALMT12/QUAC1
| Supramolecule | Name: GmALMT12/QUAC1 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: GmALMT12/QUAC1
| Macromolecule | Name: GmALMT12/QUAC1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 60.321969 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVPKVYAGQE MAMVENENCI MNGKWKKRVH VFGERVMRFP NKAWQTTWKV GREDPRRLIH AFKVGLSLTL ASLLYLLEPL FKGIGQSAI WAVMTVVVVL EFTAGATLCK GLNRGLGTLL AGLLAFLVGY IANASDRVSQ AIIIGAAVFF IGALATYMRF I PYIKKNYD ...String: MVPKVYAGQE MAMVENENCI MNGKWKKRVH VFGERVMRFP NKAWQTTWKV GREDPRRLIH AFKVGLSLTL ASLLYLLEPL FKGIGQSAI WAVMTVVVVL EFTAGATLCK GLNRGLGTLL AGLLAFLVGY IANASDRVSQ AIIIGAAVFF IGALATYMRF I PYIKKNYD YGLVIFLLTF NLITVSSYRL ENVLKIAHDR VYTIAIGCAV CLLMSLLVFP NWSGEDLHNS TVYKLEGLAK SI EACVNEY FYGEIEGSGY MKLSEDPIYK GYKAVLDSKS IDETLALHAS WEPRHSRYCH RFPWQQYVKV GAVLRQFGYT VVA LHGCLR TEIQTPRSVR AMFKDPCIRL AAEVSKVLIE LSNSIRNRRH CSPEILSDHL HEALQDLNTA IKSQPRLFLG PKHR HNQAT NMLKIAAAQV GQERHGKTSL SSVKTDSSAL LEWKTKRVSA EQTKESERKS LRPQLSKIAI TSLEFSEALP FAAFA SLLV ETVAKLDLVI EEVEELGRLA CFKEFIPGDE FVVTCQEPRV DVSQNHLPSH GVD UniProtKB: Aluminum-activated malate transporter 12 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 6189 / Average exposure time: 8.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7w6k: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)