+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31501 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | GHRP-6-bound ghrelin receptor in complex with Gq | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ghrelin / GPCR / Gq / MEMBRANE PROTEIN / GHRP-6 | |||||||||
| Function / homology |  Function and homology information Function and homology informationOlfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 ...Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / protein-containing complex binding / signal transduction / extracellular exosome / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) / Homo sapiens (human) / synthetic construct (others) /   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Wang Y / Zhuang Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Molecular recognition of an acyl-peptide hormone and activation of ghrelin receptor. Authors: Yue Wang / Shimeng Guo / Youwen Zhuang / Ying Yun / Peiyu Xu / Xinheng He / Jia Guo / Wanchao Yin / H Eric Xu / Xin Xie / Yi Jiang /  Abstract: Ghrelin, also called "the hunger hormone", is a gastric peptide hormone that regulates food intake, body weight, as well as taste sensation, reward, cognition, learning and memory. One unique feature ...Ghrelin, also called "the hunger hormone", is a gastric peptide hormone that regulates food intake, body weight, as well as taste sensation, reward, cognition, learning and memory. One unique feature of ghrelin is its acylation, primarily with an octanoic acid, which is essential for its binding and activation of the ghrelin receptor, a G protein-coupled receptor. The multifaceted roles of ghrelin make ghrelin receptor a highly attractive drug target for growth retardation, obesity, and metabolic disorders. Here we present two cryo-electron microscopy structures of G-coupled ghrelin receptor bound to ghrelin and a synthetic agonist, GHRP-6. Analysis of these two structures reveals a unique binding pocket for the octanoyl group, which guides the correct positioning of the peptide to initiate the receptor activation. Together with mutational and functional data, our structures define the rules for recognition of the acylated peptide hormone and activation of ghrelin receptor, and provide structural templates to facilitate drug design targeting ghrelin receptor. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31501.map.gz emd_31501.map.gz | 19.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31501-v30.xml emd-31501-v30.xml emd-31501.xml emd-31501.xml | 27.7 KB 27.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31501.png emd_31501.png | 53.9 KB | ||
| Filedesc metadata |  emd-31501.cif.gz emd-31501.cif.gz | 8.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31501 http://ftp.pdbj.org/pub/emdb/structures/EMD-31501 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31501 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31501 | HTTPS FTP |
-Validation report
| Summary document |  emd_31501_validation.pdf.gz emd_31501_validation.pdf.gz | 420.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31501_full_validation.pdf.gz emd_31501_full_validation.pdf.gz | 420.5 KB | Display | |
| Data in XML |  emd_31501_validation.xml.gz emd_31501_validation.xml.gz | 5.5 KB | Display | |
| Data in CIF |  emd_31501_validation.cif.gz emd_31501_validation.cif.gz | 6.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31501 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31501 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31501 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31501 | HTTPS FTP |
-Related structure data
| Related structure data |  7f9zMC  7f9yC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31501.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31501.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
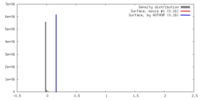
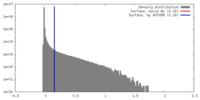
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : GHRP-6-bound ghrelin receptor in complex with Gq
| Entire | Name: GHRP-6-bound ghrelin receptor in complex with Gq |
|---|---|
| Components |
|
-Supramolecule #1: GHRP-6-bound ghrelin receptor in complex with Gq
| Supramolecule | Name: GHRP-6-bound ghrelin receptor in complex with Gq / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Engineered G-alpha-q
| Macromolecule | Name: Engineered G-alpha-q / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.855578 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMGCTLSAED KAAVERSKMI EKQLQKDKQV YRRTLRLLLL GADNSGKSTI VKQMRIYHVN GYSEEECKQY KAVVYSNTIQ SIIAIIRAM GRLKIDFGDS ARADDARQLF VLAGAAEEGF MTAELAGVIK RLWKDSGVQA CFNRSREYQL NDSAAYYLND L DRIAQPNY ...String: MMGCTLSAED KAAVERSKMI EKQLQKDKQV YRRTLRLLLL GADNSGKSTI VKQMRIYHVN GYSEEECKQY KAVVYSNTIQ SIIAIIRAM GRLKIDFGDS ARADDARQLF VLAGAAEEGF MTAELAGVIK RLWKDSGVQA CFNRSREYQL NDSAAYYLND L DRIAQPNY IPTQQDVLRT RVKTSGIFET KFQVDKVNFH MFDVGAQRDE RRKWIQCFND VTAIIFVVDS SDYNRLQEAL ND FKSIWNN RWLRTISVIL FLNKQDLLAE KVLAGKSKIE DYFPEFARYT TPEDATPEPG EDPRVTRAKY FIRKEFVDIS TAS GDGRHI CYPHFTCSVD TENARRIFND CKDIILQMNL REYNLV |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.226992 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWNG SSGGGGSGGG GSSGVSGWRL FKKIS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE
| Macromolecule | Name: 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 873.014 Da |
| Sequence | String: (DHI)(DTR)(DAL)(DTR)(DPN)(DLY)(NH2) |
-Macromolecule #4: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.736527 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAVQ LVESGGGLVQ PGGSRKLSCS ASGFAFSSFG MHWVRQAPEK GLEWVAYIS SGSGTIYYAD TVKGRFTISR DDPKNTLFLQ MTSLRSEDTA MYYCVRSIYY YGSSPFDFWG QGTTLTVSAG G GGSGGGGS ...String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAVQ LVESGGGLVQ PGGSRKLSCS ASGFAFSSFG MHWVRQAPEK GLEWVAYIS SGSGTIYYAD TVKGRFTISR DDPKNTLFLQ MTSLRSEDTA MYYCVRSIYY YGSSPFDFWG QGTTLTVSAG G GGSGGGGS GGGGSSDIVM TQATSSVPVT PGESVSISCR SSKSLLHSNG NTYLYWFLQR PGQSPQLLIY RMSNLASGVP DR FSGSGSG TAFTLTISRL EAEDVGVYYC MQHLEYPLTF GAGTKLELLE ENLYFQGASH HHHHHHH |
-Macromolecule #5: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #6: Nb35
| Macromolecule | Name: Nb35 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.343019 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAQVQLQESG GGLVQPGGSL RLSCAASGFT FSNYKMNWVR QAPGKGLEWV SDISQSGASI SYTGSVKGRF TISRDNAKNT LYLQMNSLK PEDTAVYYCA RCPAPFTRDC FDVTSTTYAY RGQGTQVTVS SHHHHHHEPE A |
-Macromolecule #7: human ghrelin receptor
| Macromolecule | Name: human ghrelin receptor / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 74.282609 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFAHHHH HHHHHHADLE DNWETLNDNL KVIEKADNAA QVKDALTKMR AAALDAQKAT PPKLEDKSPD SPEMKDFRH GFDILVGQID DALKLANEGK VKEAQAAAEQ LKTTRNAYIQ KYLMWNATPS EEPGFNLTLA DLDWDASPGN D SLGDELLQ ...String: MKTIIALSYI FCLVFAHHHH HHHHHHADLE DNWETLNDNL KVIEKADNAA QVKDALTKMR AAALDAQKAT PPKLEDKSPD SPEMKDFRH GFDILVGQID DALKLANEGK VKEAQAAAEQ LKTTRNAYIQ KYLMWNATPS EEPGFNLTLA DLDWDASPGN D SLGDELLQ LFPAPLLAGV TATCVALFVV GIAGNLLTML VVSRFRELRT TTNLYLSSMA FSDLLIFLCM PLDLVRLWQY RP WNFGDLL CKLFQFVSES CTYATVLTIT ALSVERYFAI CFPLRAKVVV TKGRVKLVIF VIWAVAFCSA GPIFVLVGVE HEN GTDPWD TNECRPTEFA VRSGLLTVMV WVSSIFFFLP VFCLTVLYSL IGRKLWRRRR GDAVVGASLR DQNHKQTVKM LAVV VFAFI LCWLPFHVGR YLFSKSFEPG SLEIAQISQY CNLVSFVLFY LSAAINPILY NIMSKKYRVA VFRLLGFEPF SQRKL STLK DESSRAWTES SINTGSAGSV FTLEDFVGDW EQTAAYNLDQ VLEQGGVSSL LQNLAVSVTP IQRIVRSGEN ALKIDI HVI IPYEGLSADQ MAQIEEVFKV VYPVDDHHFK VILPYGTLVI DGVTPNMLNY FGRPYEGIAV FDGKKITVTG TLWNGNK II DERLITPDGS MLFRVTINS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)