[English] 日本語
 Yorodumi
Yorodumi- EMDB-31443: Lysophospholipid acyltransferase LPCAT3 in a complex with Arachid... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31443 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Lysophospholipid acyltransferase LPCAT3 in a complex with Arachidonoyl-CoA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Lysophospholipid Acyltransferase / LPCAT3 / membrane-bound O-acyltransferase / cryo-EM / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationAcyl chain remodelling of PC / Acyl chain remodelling of PS / Acyl chain remodelling of PE / lysophospholipid acyltransferase activity / 1-acylglycerophosphocholine O-acyltransferase activity / lipid modification / phosphatidylcholine biosynthetic process / acyltransferase activity / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.49 Å | |||||||||
 Authors Authors | Zhang Q / Yao D | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3. Authors: Qing Zhang / Deqiang Yao / Bing Rao / Liyan Jian / Yang Chen / Kexin Hu / Ying Xia / Shaobai Li / Yafeng Shen / An Qin / Jie Zhao / Lu Zhou / Ming Lei / Xian-Cheng Jiang / Yu Cao /   Abstract: As the major component of cell membranes, phosphatidylcholine (PC) is synthesized de novo in the Kennedy pathway and then undergoes extensive deacylation-reacylation remodeling via Lands' cycle. The ...As the major component of cell membranes, phosphatidylcholine (PC) is synthesized de novo in the Kennedy pathway and then undergoes extensive deacylation-reacylation remodeling via Lands' cycle. The re-acylation is catalyzed by lysophosphatidylcholine acyltransferase (LPCAT) and among the four LPCAT members in human, the LPCAT3 preferentially introduces polyunsaturated acyl onto the sn-2 position of lysophosphatidylcholine, thereby modulating the membrane fluidity and membrane protein functions therein. Combining the x-ray crystallography and the cryo-electron microscopy, we determined the structures of LPCAT3 in apo-, acyl donor-bound, and acyl receptor-bound states. A reaction chamber was revealed in the LPCAT3 structure where the lysophosphatidylcholine and arachidonoyl-CoA were positioned in two tunnels connected near to the catalytic center. A side pocket was found expanding the tunnel for the arachidonoyl CoA and holding the main body of arachidonoyl. The structural and functional analysis provides the basis for the re-acylation of lysophosphatidylcholine and the substrate preference during the reactions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31443.map.gz emd_31443.map.gz | 72.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31443-v30.xml emd-31443-v30.xml emd-31443.xml emd-31443.xml | 10.7 KB 10.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31443.png emd_31443.png | 68.5 KB | ||
| Filedesc metadata |  emd-31443.cif.gz emd-31443.cif.gz | 5.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31443 http://ftp.pdbj.org/pub/emdb/structures/EMD-31443 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31443 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31443 | HTTPS FTP |
-Validation report
| Summary document |  emd_31443_validation.pdf.gz emd_31443_validation.pdf.gz | 472.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31443_full_validation.pdf.gz emd_31443_full_validation.pdf.gz | 472.4 KB | Display | |
| Data in XML |  emd_31443_validation.xml.gz emd_31443_validation.xml.gz | 6.2 KB | Display | |
| Data in CIF |  emd_31443_validation.cif.gz emd_31443_validation.cif.gz | 7.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31443 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31443 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31443 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31443 | HTTPS FTP |
-Related structure data
| Related structure data |  7f40MC  7ewtC  7f3xC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31443.map.gz / Format: CCP4 / Size: 76.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31443.map.gz / Format: CCP4 / Size: 76.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Lysophospholipid acyltransferase 5
| Entire | Name: Lysophospholipid acyltransferase 5 |
|---|---|
| Components |
|
-Supramolecule #1: Lysophospholipid acyltransferase 5
| Supramolecule | Name: Lysophospholipid acyltransferase 5 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: LPCAT3
| Macromolecule | Name: LPCAT3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 57.26507 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGAERDESGA GAAVLVPLLG FGSNRPAGPA EKMAVAGSGW SLARVAEALG SSEQALRLIV SILMGYPFAL FQRYFLFQKE TYLIHLYNV FTGLSIAYFN FGMQFFHSLL CVLIQFLILR LMGRTVTAVF TTFVFQMTYL MAGYYFTATE HYDIKWTMPH C VLTLKLIG ...String: MGAERDESGA GAAVLVPLLG FGSNRPAGPA EKMAVAGSGW SLARVAEALG SSEQALRLIV SILMGYPFAL FQRYFLFQKE TYLIHLYNV FTGLSIAYFN FGMQFFHSLL CVLIQFLILR LMGRTVTAVF TTFVFQMTYL MAGYYFTATE HYDIKWTMPH C VLTLKLIG LAIDYYDGGK DPELLTPEQR RFAVRGVPTL LEVSGFSYFY GAFMVGPQFS MTDYQKLAKG EMTDVPGQRP NS FVPALKR LSLGLLFLVT YTLSSPYISE EYLISDDYME KPFWFRCGYI LVWGKIILYK YVTCWLVTEG VCILVGLGYN GND QNGKPV WDACANMKVW LYETTPLFTG TIASFNINTN AWVARYVFKR LKFLGNKLLS QALALFFLAI WHGLHSGYLV CFQM ELLIV IVERQVINLV RDSPTLSTLA SITALQPIFY VLQQTNHWMF MGYSLVPFCL FTWDKWMKVY KSIYFLGHVL FFTLL LVLP YIRKLLVPRK EKLKKAE UniProtKB: Lysophosphatidylcholine acyltransferase 3 |
-Macromolecule #2: S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidany...
| Macromolecule | Name: S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino] ...Name: S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenethioate type: ligand / ID: 2 / Number of copies: 2 / Formula: 3IX |
|---|---|
| Molecular weight | Theoretical: 1.053986 KDa |
| Chemical component information | 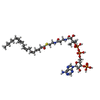 ChemComp-3IX: |
-Macromolecule #3: 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 3 / Number of copies: 2 / Formula: PCW |
|---|---|
| Molecular weight | Theoretical: 787.121 Da |
| Chemical component information |  ChemComp-PCW: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.49 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 292613 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















