+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31440 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human TMEM120A in the CoASH-bound state | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Human / membrane protein / COA | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationcoenzyme A binding / protein heterooligomerization / nuclear inner membrane / fat cell differentiation / monoatomic ion channel activity / detection of mechanical stimulus involved in sensory perception of pain / antiviral innate immune response / protein homooligomerization / monoatomic ion transmembrane transport / endoplasmic reticulum ...coenzyme A binding / protein heterooligomerization / nuclear inner membrane / fat cell differentiation / monoatomic ion channel activity / detection of mechanical stimulus involved in sensory perception of pain / antiviral innate immune response / protein homooligomerization / monoatomic ion transmembrane transport / endoplasmic reticulum / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
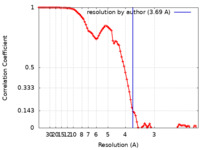
| Method | single particle reconstruction / cryo EM / Resolution: 3.69 Å | ||||||||||||
 Authors Authors | Song DF / Rong Y | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: TMEM120A contains a specific coenzyme A-binding site and might not mediate poking- or stretch-induced channel activities in cells. Authors: Yao Rong / Jinghui Jiang / Yiwei Gao / Jianli Guo / Danfeng Song / Wenhao Liu / Mingmin Zhang / Yan Zhao / Bailong Xiao / Zhenfeng Liu /  Abstract: TMEM120A, a member of the transmembrane protein 120 (TMEM120) family, has a pivotal function in adipocyte differentiation and metabolism, and may also contribute to sensing mechanical pain by ...TMEM120A, a member of the transmembrane protein 120 (TMEM120) family, has a pivotal function in adipocyte differentiation and metabolism, and may also contribute to sensing mechanical pain by functioning as an ion channel named TACAN. Here we report that expression of TMEM120A is not sufficient in mediating poking- or stretch-induced currents in cells and have solved cryo-electron microscopy (cryo-EM) structures of human TMEM120A (TMEM120A) in complex with an endogenous metabolic cofactor (coenzyme A, CoASH) and in the apo form. TMEM120A forms a symmetrical homodimer with each monomer containing an amino-terminal coiled-coil motif followed by a transmembrane domain with six membrane-spanning helices. Within the transmembrane domain, a CoASH molecule is hosted in a deep cavity and forms specific interactions with nearby amino acid residues. Mutation of a central tryptophan residue involved in binding CoASH dramatically reduced the binding affinity of TMEM120A with CoASH. TMEM120A exhibits distinct conformations at the states with or without CoASH bound. Our results suggest that TMEM120A may have alternative functional roles potentially involved in CoASH transport, sensing, or metabolism. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31440.map.gz emd_31440.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31440-v30.xml emd-31440-v30.xml emd-31440.xml emd-31440.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31440_fsc.xml emd_31440_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_31440.png emd_31440.png | 85.1 KB | ||
| Filedesc metadata |  emd-31440.cif.gz emd-31440.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31440 http://ftp.pdbj.org/pub/emdb/structures/EMD-31440 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31440 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31440 | HTTPS FTP |
-Validation report
| Summary document |  emd_31440_validation.pdf.gz emd_31440_validation.pdf.gz | 475.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31440_full_validation.pdf.gz emd_31440_full_validation.pdf.gz | 474.7 KB | Display | |
| Data in XML |  emd_31440_validation.xml.gz emd_31440_validation.xml.gz | 11 KB | Display | |
| Data in CIF |  emd_31440_validation.cif.gz emd_31440_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31440 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31440 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31440 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31440 | HTTPS FTP |
-Related structure data
| Related structure data |  7f3tMC  7f3uC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31440.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31440.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : transmembrane protein 120A
| Entire | Name: transmembrane protein 120A |
|---|---|
| Components |
|
-Supramolecule #1: transmembrane protein 120A
| Supramolecule | Name: transmembrane protein 120A / type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Transmembrane protein 120A
| Macromolecule | Name: Transmembrane protein 120A / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 44.843578 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDYKDDDDKW SHPQFEKHHH HHHHHWSHPQ FEKQPPPPGP LGDCLRDWED LQQDFQNIQE THRLYRLKLE ELTKLQNNCT SSITRQKKR LQELALALKK CKPSLPAEAE GAAQELENQM KERQGLFFDM EAYLPKKNGL YLSLVLGNVN VTLLSKQAKF A YKDEYEKF ...String: MDYKDDDDKW SHPQFEKHHH HHHHHWSHPQ FEKQPPPPGP LGDCLRDWED LQQDFQNIQE THRLYRLKLE ELTKLQNNCT SSITRQKKR LQELALALKK CKPSLPAEAE GAAQELENQM KERQGLFFDM EAYLPKKNGL YLSLVLGNVN VTLLSKQAKF A YKDEYEKF KLYLTIILIL ISFTCRFLLN SRVTDAAFNF LLVWYYCTLT IRESILINNG SRIKGWWVFH HYVSTFLSGV ML TWPDGLM YQKFRNQFLS FSMYQSFVQF LQYYYQSGCL YRLRALGERH TMDLTVEGFQ SWMWRGLTFL LPFLFFGHFW QLF NALTLF NLAQDPQCKE WQVLMCGFPF LLLFLGNFFT TLRVVHHKFH SQRHGSKKD UniProtKB: Transmembrane protein 120A |
-Macromolecule #2: COENZYME A
| Macromolecule | Name: COENZYME A / type: ligand / ID: 2 / Number of copies: 2 / Formula: COA |
|---|---|
| Molecular weight | Theoretical: 767.534 Da |
| Chemical component information |  ChemComp-COA: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | The protein is reconstituted in lipid nanodiscs |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Number real images: 8148 / Average exposure time: 1.875 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-7f3t: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)