+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31004 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Heme arrested off-pathway oligomer of alpha-synuclein | |||||||||
 Map data Map data | It is a cryo-EM map of alfa-syneuclein tetramer in presence of heme. alfa-syneuclein monomer was incubated with heme molecule to prepare the sample. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
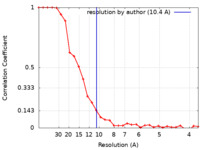
| Method | single particle reconstruction / cryo EM / Resolution: 10.4 Å | |||||||||
 Authors Authors | Dey S / Sil P / Paul S | |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2021 Journal: Commun Biol / Year: 2021Title: Conformational distortion in a fibril-forming oligomer arrests alpha-Synuclein fibrillation and minimizes its toxic effects. Authors: Ritobrita Chakraborty / Sandip Dey / Pallabi Sil / Simanta Sarani Paul / Dipita Bhattacharyya / Anirban Bhunia / Jayati Sengupta / Krishnananda Chattopadhyay /   Abstract: The fibrillation pathway of alpha-Synuclein, the causative protein of Parkinson's disease, encompasses transient, heterogeneous oligomeric forms whose structural understanding and link to toxicity ...The fibrillation pathway of alpha-Synuclein, the causative protein of Parkinson's disease, encompasses transient, heterogeneous oligomeric forms whose structural understanding and link to toxicity are not yet understood. We report that the addition of the physiologically-available small molecule heme at a sub-stoichiometric ratio to either monomeric or aggregated α-Syn, targets a His50 residue critical for fibril-formation and stabilizes the structurally-heterogeneous populations of aggregates into a minimally-toxic oligomeric state. Cryo-EM 3D reconstruction revealed a 'mace'-shaped structure of this monodisperse population of oligomers, which is comparable to a solid-state NMR Greek key-like motif (where the core residues are arranged in parallel in-register sheets with a Greek key topology at the C terminus) that forms the fundamental unit/kernel of protofilaments. Further structural analyses suggest that heme binding induces a distortion in the Greek key-like architecture of the mace oligomers, which impairs their further appending into protofilaments and fibrils. Additionally, our study reports a novel mechanism of prevention as well as reclamation of amyloid fibril formation by blocking an inter-protofilament His50 residue using a small molecule. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31004.map.gz emd_31004.map.gz | 1.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31004-v30.xml emd-31004-v30.xml emd-31004.xml emd-31004.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31004_fsc.xml emd_31004_fsc.xml | 3.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_31004.png emd_31004.png | 132.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31004 http://ftp.pdbj.org/pub/emdb/structures/EMD-31004 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31004 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31004 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31004.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31004.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | It is a cryo-EM map of alfa-syneuclein tetramer in presence of heme. alfa-syneuclein monomer was incubated with heme molecule to prepare the sample. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.89 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Heme arrested off-pathway oligomer of alfa-syneuclein
| Entire | Name: Heme arrested off-pathway oligomer of alfa-syneuclein |
|---|---|
| Components |
|
-Supramolecule #1: Heme arrested off-pathway oligomer of alfa-syneuclein
| Supramolecule | Name: Heme arrested off-pathway oligomer of alfa-syneuclein / type: complex / ID: 1 / Parent: 0 Details: To generate off-pathway oligomer, alfa-syneuclein was incubated with heme in shaking (120 rpm)condition for 22 hrs at 37C. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: brain / Tissue: brain tissue / Location in cell: Cytoplasm Homo sapiens (human) / Organ: brain / Tissue: brain tissue / Location in cell: Cytoplasm |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: sodium phosphate buffer, pH 7.4 |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Number grids imaged: 2 / Average exposure time: 2.0 sec. / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION |
| Sample stage | Specimen holder model: GATAN 910 MULTI-SPECIMEN SINGLE TILT CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)