[English] 日本語
 Yorodumi
Yorodumi- EMDB-30620: Structure of human soluble guanylate cyclase in the cinciguat-bou... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30620 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human soluble guanylate cyclase in the cinciguat-bound inactive state | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | soluble guanylate cyclase / SIGNALING PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationretrograde trans-synaptic signaling by nitric oxide, modulating synaptic transmission / cytidylate cyclase activity / guanylate cyclase complex, soluble / guanylate cyclase / cGMP biosynthetic process / guanylate cyclase activity / presynaptic active zone cytoplasmic component / response to oxygen levels / nitric oxide binding / relaxation of vascular associated smooth muscle ...retrograde trans-synaptic signaling by nitric oxide, modulating synaptic transmission / cytidylate cyclase activity / guanylate cyclase complex, soluble / guanylate cyclase / cGMP biosynthetic process / guanylate cyclase activity / presynaptic active zone cytoplasmic component / response to oxygen levels / nitric oxide binding / relaxation of vascular associated smooth muscle / Nitric oxide stimulates guanylate cyclase / adenylate cyclase activity / blood circulation / : / positive regulation of nitric oxide mediated signal transduction / nitric oxide mediated signal transduction / Smooth Muscle Contraction / nitric oxide-cGMP-mediated signaling / cellular response to nitric oxide / Hsp90 protein binding / GABA-ergic synapse / regulation of blood pressure / signaling receptor activity / heme binding / GTP binding / protein-containing complex binding / glutamatergic synapse / metal ion binding / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||||||||
 Authors Authors | Chen L / Liu R | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Activation mechanism of human soluble guanylate cyclase by stimulators and activators. Authors: Rui Liu / Yunlu Kang / Lei Chen /  Abstract: Soluble guanylate cyclase (sGC) is the receptor for nitric oxide (NO) in human. It is an important validated drug target for cardiovascular diseases. sGC can be pharmacologically activated by ...Soluble guanylate cyclase (sGC) is the receptor for nitric oxide (NO) in human. It is an important validated drug target for cardiovascular diseases. sGC can be pharmacologically activated by stimulators and activators. However, the detailed structural mechanisms, through which sGC is recognized and positively modulated by these drugs at high spacial resolution, are poorly understood. Here, we present cryo-electron microscopy structures of human sGC in complex with NO and sGC stimulators, YC-1 and riociguat, and also in complex with the activator cinaciguat. These structures uncover the molecular details of how stimulators interact with residues from both β H-NOX and CC domains, to stabilize sGC in the extended active conformation. In contrast, cinaciguat occupies the haem pocket in the β H-NOX domain and sGC shows both inactive and active conformations. These structures suggest a converged mechanism of sGC activation by pharmacological compounds. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30620.map.gz emd_30620.map.gz | 2.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30620-v30.xml emd-30620-v30.xml emd-30620.xml emd-30620.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30620.png emd_30620.png | 53.8 KB | ||
| Filedesc metadata |  emd-30620.cif.gz emd-30620.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30620 http://ftp.pdbj.org/pub/emdb/structures/EMD-30620 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30620 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30620 | HTTPS FTP |
-Validation report
| Summary document |  emd_30620_validation.pdf.gz emd_30620_validation.pdf.gz | 359.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30620_full_validation.pdf.gz emd_30620_full_validation.pdf.gz | 359.4 KB | Display | |
| Data in XML |  emd_30620_validation.xml.gz emd_30620_validation.xml.gz | 5.8 KB | Display | |
| Data in CIF |  emd_30620_validation.cif.gz emd_30620_validation.cif.gz | 6.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30620 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30620 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30620 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30620 | HTTPS FTP |
-Related structure data
| Related structure data |  7d9tMC  7d9rC  7d9sC  7d9uC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30620.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30620.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human soluble guanylate cyclase
| Entire | Name: human soluble guanylate cyclase |
|---|---|
| Components |
|
-Supramolecule #1: human soluble guanylate cyclase
| Supramolecule | Name: human soluble guanylate cyclase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanylate cyclase soluble subunit alpha-1
| Macromolecule | Name: Guanylate cyclase soluble subunit alpha-1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: guanylate cyclase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 77.566484 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFCTKLKDLK ITGECPFSLL APGQVPNESS EEAAGSSESC KATMPICQDI PEKNIQESLP QRKTSRSRVY LHTLAESICK LIFPEFERL NVALQRTLAK HKIKESRKSL EREDFEKTIA EQAVAAGVPV EVIKESLGEE VFKICYEEDE NILGVVGGTL K DFLNSFST ...String: MFCTKLKDLK ITGECPFSLL APGQVPNESS EEAAGSSESC KATMPICQDI PEKNIQESLP QRKTSRSRVY LHTLAESICK LIFPEFERL NVALQRTLAK HKIKESRKSL EREDFEKTIA EQAVAAGVPV EVIKESLGEE VFKICYEEDE NILGVVGGTL K DFLNSFST LLKQSSHCQE AGKRGRLEDA SILCLDKEDD FLHVYYFFPK RTTSLILPGI IKAAAHVLYE TEVEVSLMPP CF HNDCSEF VNQPYLLYSV HMKSTKPSLS PSKPQSSLVI PTSLFCKTFP FHFMFDKDMT ILQFGNGIRR LMNRRDFQGK PNF EEYFEI LTPKINQTFS GIMTMLNMQF VVRVRRWDNS VKKSSRVMDL KGQMIYIVES SAILFLGSPC VDRLEDFTGR GLYL SDIPI HNALRDVVLI GEQARAQDGL KKRLGKLKAT LEQAHQALEE EKKKTVDLLC SIFPCEVAQQ LWQGQVVQAK KFSNV TMLF SDIVGFTAIC SQCSPLQVIT MLNALYTRFD QQCGELDVYK VETIGDAYCV AGGLHKESDT HAVQIALMAV KMMELS DEV MSPHGEPIKM RIGLHSGSVF AGVVGVKMPR YCLFGNNVTL ANKFESCSVP RKINVSPTTY RLLKDCPGFV FTPRSRE EL PPNFPSEIPG ICHFLDAYQQ GTNSKPCFQK KDVEDGNANF LGKASGID UniProtKB: Guanylate cyclase soluble subunit alpha-1 |
-Macromolecule #2: Guanylate cyclase soluble subunit beta-1
| Macromolecule | Name: Guanylate cyclase soluble subunit beta-1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: guanylate cyclase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 70.59932 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MYGFVNHALE LLVIRNYGPE VWEDIKKEAQ LDEEGQFLVR IIYDDSKTYD LVAAASKVLN LNAGEILQMF GKMFFVFCQE SGYDTILRV LGSNVREFLQ NLDALHDHLA TIYPGMRAPS FRCTDAEKGK GLILHYYSER EGLQDIVIGI IKTVAQQIHG T EIDMKVIQ ...String: MYGFVNHALE LLVIRNYGPE VWEDIKKEAQ LDEEGQFLVR IIYDDSKTYD LVAAASKVLN LNAGEILQMF GKMFFVFCQE SGYDTILRV LGSNVREFLQ NLDALHDHLA TIYPGMRAPS FRCTDAEKGK GLILHYYSER EGLQDIVIGI IKTVAQQIHG T EIDMKVIQ QRNEECDHTQ FLIEEKESKE EDFYEDLDRF EENGTQESRI SPYTFCKAFP FHIIFDRDLV VTQCGNAIYR VL PQLQPGN CSLLSVFSLV RPHIDISFHG ILSHINTVFV LRSKEGLLDV EKLECEDELT GTEISCLRLK GQMIYLPEAD SIL FLCSPS VMNLDDLTRR GLYLSDIPLH DATRDLVLLG EQFREEYKLT QELEILTDRL QLTLRALEDE KKKTDTLLYS VLPP SVANE LRHKRPVPAK RYDNVTILFS GIVGFNAFCS KHASGEGAMK IVNLLNDLYT RFDTLTDSRK NPFVYKVETV GDKYM TVSG LPEPCIHHAR SICHLALDMM EIAGQVQVDG ESVQITIGIH TGEVVTGVIG QRMPRYCLFG NTVNLTSRTE TTGEKG KIN VSEYTYRCLM SPENSDPQFH LEHRGPVSMK GKKEPMQVWF LSRKNTGTEE TKQDDD UniProtKB: Guanylate cyclase soluble subunit beta-1 |
-Macromolecule #3: 4-({(4-carboxybutyl)[2-(2-{[4-(2-phenylethyl)benzyl]oxy}phenyl)et...
| Macromolecule | Name: 4-({(4-carboxybutyl)[2-(2-{[4-(2-phenylethyl)benzyl]oxy}phenyl)ethyl]amino}methyl)benzoic acid type: ligand / ID: 3 / Number of copies: 1 / Formula: Z90 |
|---|---|
| Molecular weight | Theoretical: 565.699 Da |
| Chemical component information | 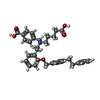 ChemComp-Z90: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.1 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 244400 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















