+Search query
-Structure paper
| Title | Activation mechanism of human soluble guanylate cyclase by stimulators and activators. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 12, Issue 1, Page 5492, Year 2021 |
| Publish date | Sep 17, 2021 |
 Authors Authors | Rui Liu / Yunlu Kang / Lei Chen /  |
| PubMed Abstract | Soluble guanylate cyclase (sGC) is the receptor for nitric oxide (NO) in human. It is an important validated drug target for cardiovascular diseases. sGC can be pharmacologically activated by ...Soluble guanylate cyclase (sGC) is the receptor for nitric oxide (NO) in human. It is an important validated drug target for cardiovascular diseases. sGC can be pharmacologically activated by stimulators and activators. However, the detailed structural mechanisms, through which sGC is recognized and positively modulated by these drugs at high spacial resolution, are poorly understood. Here, we present cryo-electron microscopy structures of human sGC in complex with NO and sGC stimulators, YC-1 and riociguat, and also in complex with the activator cinaciguat. These structures uncover the molecular details of how stimulators interact with residues from both β H-NOX and CC domains, to stabilize sGC in the extended active conformation. In contrast, cinaciguat occupies the haem pocket in the β H-NOX domain and sGC shows both inactive and active conformations. These structures suggest a converged mechanism of sGC activation by pharmacological compounds. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:34535643 / PubMed:34535643 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.7 - 4.1 Å |
| Structure data | EMDB-30618, PDB-7d9r: EMDB-30619, PDB-7d9s: EMDB-30620, PDB-7d9t: EMDB-30621, PDB-7d9u: |
| Chemicals |  ChemComp-G2P:  ChemComp-MG:  ChemComp-HEM: 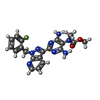 ChemComp-GZO: 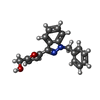 ChemComp-YC1: 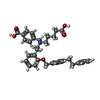 ChemComp-Z90: |
| Source |
|
 Keywords Keywords | SIGNALING PROTEIN / soluble guanylate cyclase |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers











 homo sapiens (human)
homo sapiens (human)