[English] 日本語
 Yorodumi
Yorodumi- EMDB-30611: Cryo-EM map of 70S ribosome in complex with peptide deformylase, ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30611 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of 70S ribosome in complex with peptide deformylase, trigger factor, and methionine aminopeptidase | ||||||||||||
 Map data Map data | Cryo-EM map of E. coli 70S ribosome in complex with peptide deformylase, trigger factor, and methionine aminopeptidase | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Escherichia coli / ribosome / nascent chain / protein biogenesis / peptide deformylase / trigger factor / methionine aminopeptidase | ||||||||||||
| Function / homology |  Function and homology information Function and homology information'de novo' cotranslational protein folding / stress response to copper ion / peptide deformylase / peptide deformylase activity / co-translational protein modification / stringent response / ornithine decarboxylase inhibitor activity / transcription antitermination factor activity, RNA binding / misfolded RNA binding / Group I intron splicing ...'de novo' cotranslational protein folding / stress response to copper ion / peptide deformylase / peptide deformylase activity / co-translational protein modification / stringent response / ornithine decarboxylase inhibitor activity / transcription antitermination factor activity, RNA binding / misfolded RNA binding / Group I intron splicing / RNA folding / protein unfolding / transcriptional attenuation / positive regulation of ribosome biogenesis / endoribonuclease inhibitor activity / RNA-binding transcription regulator activity / translational termination / negative regulation of cytoplasmic translation / four-way junction DNA binding / DnaA-L2 complex / : / translation repressor activity / negative regulation of translational initiation / regulation of mRNA stability / negative regulation of DNA-templated DNA replication initiation / protein folding chaperone / mRNA regulatory element binding translation repressor activity / assembly of large subunit precursor of preribosome / positive regulation of RNA splicing / ribosome assembly / peptidyl-prolyl cis-trans isomerase activity / transcription elongation factor complex / regulation of DNA-templated transcription elongation / cytosolic ribosome assembly / response to reactive oxygen species / DNA endonuclease activity / transcription antitermination / RNA polymerase II CTD heptapeptide repeat P3 isomerase activity / RNA polymerase II CTD heptapeptide repeat P6 isomerase activity / peptidylprolyl isomerase / regulation of cell growth / DNA-templated transcription termination / response to radiation / ferrous iron binding / maintenance of translational fidelity / mRNA 5'-UTR binding / protein transport / regulation of translation / ribosome biogenesis / large ribosomal subunit / transferase activity / ribosome binding / ribosomal small subunit biogenesis / ribosomal small subunit assembly / response to heat / small ribosomal subunit / 5S rRNA binding / ribosomal large subunit assembly / cytosolic small ribosomal subunit / large ribosomal subunit rRNA binding / small ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / hydrolase activity / negative regulation of translation / rRNA binding / ribosome / structural constituent of ribosome / translation / cell division / response to antibiotic / negative regulation of DNA-templated transcription / mRNA binding / DNA binding / RNA binding / zinc ion binding / identical protein binding / membrane / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |   | ||||||||||||
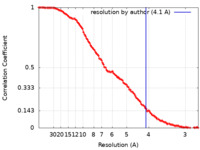
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | ||||||||||||
 Authors Authors | Akbar S / Bhakta S | ||||||||||||
| Funding support |  India, 3 items India, 3 items
| ||||||||||||
 Citation Citation |  Journal: Structure / Year: 2021 Journal: Structure / Year: 2021Title: Structural insights into the interplay of protein biogenesis factors with the 70S ribosome. Authors: Shirin Akbar / Sayan Bhakta / Jayati Sengupta /  Abstract: Bacterial co-translational N-terminal methionine excision, an early event of nascent polypeptide chain processing, is mediated by two enzymes: peptide deformylase (PDF) and methionine aminopeptidase ...Bacterial co-translational N-terminal methionine excision, an early event of nascent polypeptide chain processing, is mediated by two enzymes: peptide deformylase (PDF) and methionine aminopeptidase (MetAP). Trigger factor (TF), the only ribosome-associated bacterial chaperone, offers co-translational chaperoning assistance. Here, we present two high-resolution cryoelectron microscopy structures of tRNA-bound E. coli ribosome complexes showing simultaneous binding of PDF and TF, in the absence (3.4 Å) and presence of MetAP (4.1 Å). These structures establish molecular details of the interactions of the factors with the ribosome, and thereby reveal the structural basis of nascent chain processing. Our results suggest that simultaneous binding of all three factors is not a functionally favorable mechanism of nascent chain processing. Strikingly, an unusual structural distortion of the 70S ribosome, potentially driven by binding of multiple copies of MetAP, is observed when MetAP is incubated with a pre-formed PDF-TF-bound ribosome complex. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30611.map.gz emd_30611.map.gz | 98.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30611-v30.xml emd-30611-v30.xml emd-30611.xml emd-30611.xml | 70.7 KB 70.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30611_fsc.xml emd_30611_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_30611.png emd_30611.png | 197.9 KB | ||
| Filedesc metadata |  emd-30611.cif.gz emd-30611.cif.gz | 14.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30611 http://ftp.pdbj.org/pub/emdb/structures/EMD-30611 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30611 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30611 | HTTPS FTP |
-Related structure data
| Related structure data |  7d80MC  7d6zC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30611.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30611.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of E. coli 70S ribosome in complex with peptide deformylase, trigger factor, and methionine aminopeptidase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : E. coli 70S ribosome in complex with enzyme peptide deformylase, ...
+Supramolecule #1: E. coli 70S ribosome in complex with enzyme peptide deformylase, ...
+Macromolecule #1: 50S ribosomal protein L34
+Macromolecule #2: 50S ribosomal protein L35
+Macromolecule #3: 50S ribosomal protein L36
+Macromolecule #4: Peptide deformylase
+Macromolecule #5: Trigger factor
+Macromolecule #6: 50S ribosomal protein L32
+Macromolecule #9: 30S ribosomal protein S2
+Macromolecule #10: 30S ribosomal protein S3
+Macromolecule #11: 30S ribosomal protein S4
+Macromolecule #12: 30S ribosomal protein S5
+Macromolecule #13: 30S ribosomal protein S6, fully modified isoform
+Macromolecule #14: 30S ribosomal protein S7
+Macromolecule #15: 30S ribosomal protein S8
+Macromolecule #16: 30S ribosomal protein S9
+Macromolecule #17: 30S ribosomal protein S10
+Macromolecule #18: 30S ribosomal protein S11
+Macromolecule #19: 30S ribosomal protein S12
+Macromolecule #20: 30S ribosomal protein S13
+Macromolecule #21: 30S ribosomal protein S14
+Macromolecule #22: 30S ribosomal protein S15
+Macromolecule #23: 30S ribosomal protein S16
+Macromolecule #24: 30S ribosomal protein S17
+Macromolecule #25: 30S ribosomal protein S18
+Macromolecule #26: 30S ribosomal protein S19
+Macromolecule #27: 30S ribosomal protein S20
+Macromolecule #28: 30S ribosomal protein S21
+Macromolecule #29: 50S ribosomal protein L23
+Macromolecule #33: 50S ribosomal protein L2
+Macromolecule #34: 50S ribosomal protein L3
+Macromolecule #35: 50S ribosomal protein L4
+Macromolecule #36: 50S ribosomal protein L5
+Macromolecule #37: 50S ribosomal protein L6
+Macromolecule #38: 50S ribosomal protein L9
+Macromolecule #39: 50S ribosomal protein L11
+Macromolecule #40: 50S ribosomal protein L13
+Macromolecule #41: 50S ribosomal protein L14
+Macromolecule #42: 50S ribosomal protein L15
+Macromolecule #43: 50S ribosomal protein L16
+Macromolecule #44: 50S ribosomal protein L17
+Macromolecule #45: 50S ribosomal protein L18
+Macromolecule #46: 50S ribosomal protein L19
+Macromolecule #47: 50S ribosomal protein L20
+Macromolecule #48: 50S ribosomal protein L21
+Macromolecule #49: 50S ribosomal protein L22
+Macromolecule #50: 50S ribosomal protein L24
+Macromolecule #51: 50S ribosomal protein L25
+Macromolecule #52: 50S ribosomal protein L27
+Macromolecule #53: 50S ribosomal protein L28
+Macromolecule #54: 50S ribosomal protein L29
+Macromolecule #55: 50S ribosomal protein L30
+Macromolecule #57: 50S ribosomal protein L33
+Macromolecule #7: 23S ribosomal RNA
+Macromolecule #8: 16S ribosomal RNA
+Macromolecule #30: E-site tRNA
+Macromolecule #31: A-site tRNA
+Macromolecule #32: 5S ribosomal RNA
+Macromolecule #56: P-site tRNA
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 32.57 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)