[English] 日本語
 Yorodumi
Yorodumi- EMDB-30479: Cryo-EM structure of Chikungunya virus in complex with Fab fragme... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30479 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Chikungunya virus in complex with Fab fragments of mAb CHK-263 (subregion around icosahedral 5-fold vertex) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CHIKV / Fab / complex / localized reconstruction / VIRUS / VIRUS-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationtogavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / viral translational frameshifting / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion attachment to host cell / host cell nucleus ...togavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / viral translational frameshifting / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion attachment to host cell / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / RNA binding / membrane Similarity search - Function | |||||||||
| Biological species |   Chikungunya virus / Chikungunya virus /  | |||||||||
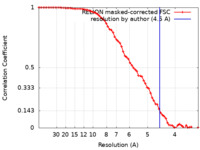
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Zhou QF / Fox JM / Earnest JT / Ng TS / Kim AS / Fibriansah G / Kostyuchenko VA / Shu B / Diamond MS / Lok SM | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Structural basis of Chikungunya virus inhibition by monoclonal antibodies. Authors: Qun Fei Zhou / Julie M Fox / James T Earnest / Thiam-Seng Ng / Arthur S Kim / Guntur Fibriansah / Victor A Kostyuchenko / Jian Shi / Bo Shu / Michael S Diamond / Shee-Mei Lok /   Abstract: Chikungunya virus (CHIKV) is an emerging viral pathogen that causes both acute and chronic debilitating arthritis. Here, we describe the functional and structural basis as to how two anti-CHIKV ...Chikungunya virus (CHIKV) is an emerging viral pathogen that causes both acute and chronic debilitating arthritis. Here, we describe the functional and structural basis as to how two anti-CHIKV monoclonal antibodies, CHK-124 and CHK-263, potently inhibit CHIKV infection in vitro and in vivo. Our in vitro studies show that CHK-124 and CHK-263 block CHIKV at multiple stages of viral infection. CHK-124 aggregates virus particles and blocks attachment. Also, due to antibody-induced virus aggregation, fusion with endosomes and egress are inhibited. CHK-263 neutralizes CHIKV infection mainly by blocking virus attachment and fusion. To determine the structural basis of neutralization, we generated cryogenic electron microscopy reconstructions of Fab:CHIKV complexes at 4- to 5-Å resolution. CHK-124 binds to the E2 domain B and overlaps with the Mxra8 receptor-binding site. CHK-263 blocks fusion by binding an epitope that spans across E1 and E2 and locks the heterodimer together, likely preventing structural rearrangements required for fusion. These results provide structural insight as to how neutralizing antibody engagement of CHIKV inhibits different stages of the viral life cycle, which could inform vaccine and therapeutic design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
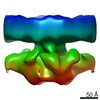
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30479.map.gz emd_30479.map.gz | 12.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30479-v30.xml emd-30479-v30.xml emd-30479.xml emd-30479.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30479_fsc.xml emd_30479_fsc.xml | 6.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_30479.png emd_30479.png | 280.3 KB | ||
| Filedesc metadata |  emd-30479.cif.gz emd-30479.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30479 http://ftp.pdbj.org/pub/emdb/structures/EMD-30479 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30479 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30479 | HTTPS FTP |
-Validation report
| Summary document |  emd_30479_validation.pdf.gz emd_30479_validation.pdf.gz | 626.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30479_full_validation.pdf.gz emd_30479_full_validation.pdf.gz | 625.8 KB | Display | |
| Data in XML |  emd_30479_validation.xml.gz emd_30479_validation.xml.gz | 9.4 KB | Display | |
| Data in CIF |  emd_30479_validation.cif.gz emd_30479_validation.cif.gz | 12.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30479 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30479 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30479 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30479 | HTTPS FTP |
-Related structure data
| Related structure data |  7cw2MC  7cvyC  7cvzC  7cw0C  7cw3C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30479.map.gz / Format: CCP4 / Size: 25.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30479.map.gz / Format: CCP4 / Size: 25.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.71 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
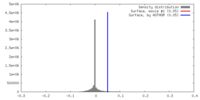
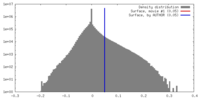
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Localized reconstruction of subregions around the icosahedral 5-f...
| Entire | Name: Localized reconstruction of subregions around the icosahedral 5-fold vertex of CHIKV:CHK-263 Fab complex |
|---|---|
| Components |
|
-Supramolecule #1: Localized reconstruction of subregions around the icosahedral 5-f...
| Supramolecule | Name: Localized reconstruction of subregions around the icosahedral 5-fold vertex of CHIKV:CHK-263 Fab complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Chikungunya virus
| Supramolecule | Name: Chikungunya virus / type: organelle_or_cellular_component / ID: 2 / Parent: 1 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Chikungunya virus Chikungunya virus |
-Supramolecule #3: Fab region of CHK-263
| Supramolecule | Name: Fab region of CHK-263 / type: organelle_or_cellular_component / ID: 3 / Parent: 1 / Macromolecule list: #4-#5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: E1 glycoprotein
| Macromolecule | Name: E1 glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Chikungunya virus Chikungunya virus |
| Molecular weight | Theoretical: 47.503016 KDa |
| Sequence | String: YEHVTVIPNT VGVPYKTLVN RPGYSPMVLE MELLSVTLEP TLSLDYITCE YKTVIPSPYV KCCGTAECKD KNLPDYSCKV FTGVYPFMW GGAYCFCDAE NTQLSEAHVE KSESCKTEFA SAYRAHTASA SAKLRVLYQG NNITVTAYAN GDHAVTVKDA K FIVGPMSS ...String: YEHVTVIPNT VGVPYKTLVN RPGYSPMVLE MELLSVTLEP TLSLDYITCE YKTVIPSPYV KCCGTAECKD KNLPDYSCKV FTGVYPFMW GGAYCFCDAE NTQLSEAHVE KSESCKTEFA SAYRAHTASA SAKLRVLYQG NNITVTAYAN GDHAVTVKDA K FIVGPMSS AWTPFDNKIV VYKGDVYNMD YPPFGAGRPG QFGDIQSRTP ESKDVYANTQ LVLQRPAAGT VHVPYSQAPS GF KYWLKER GASLQHTAPF GCQIATNPVR AVNCAVGNMP ISIDIPEAAF TRVVDAPSLT DMSCEVPACT HSSDFGGVAI IKY AASKKG KCAVHSMTNA VTIREAEIEV EGNSQLQISF STALASAEFR VQVCSTQVHC AAECHPPKRT TVYYPASHTT LGVQ DISAT AMSWVQKITG GVGLVVAVAA LILIVVLCVS FSRH |
-Macromolecule #2: E2 glycoprotein
| Macromolecule | Name: E2 glycoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 15 / Enantiomer: LEVO / EC number: togavirin |
|---|---|
| Source (natural) | Organism:   Chikungunya virus Chikungunya virus |
| Molecular weight | Theoretical: 46.904559 KDa |
| Sequence | String: NFNVYKATRP YLAHCPDCGE GHSCHSPVAL ERIRNEATDG TLKIQVSLQI GIKTDDSHDW TKLRYMDNHM PADAERAGLF VRTSAPCTI TGTIGHFILA RCPKGETLTV GFTDSRKISH SCTHPFHHDP PVIGREKFHS RPQHGKELPC STYVQSTAAT T EEIEVHMP ...String: NFNVYKATRP YLAHCPDCGE GHSCHSPVAL ERIRNEATDG TLKIQVSLQI GIKTDDSHDW TKLRYMDNHM PADAERAGLF VRTSAPCTI TGTIGHFILA RCPKGETLTV GFTDSRKISH SCTHPFHHDP PVIGREKFHS RPQHGKELPC STYVQSTAAT T EEIEVHMP PDTPDHTLMS QQSGNVKITV NGQTVRYKCN CGGSNEGLTT TDKVINNCKV DQCHAAVTNH KKWQYNSPLV PR NAELGDR KGKIHIPFPL ANVTCRVPKA RNPTVTYGKN QVIMLLYPDH PTLLSYRNMG EEPNYQEEWV MHKKEVVLTV PTE GLEVTW GNNEPYKYWP QLSTNGTAHG HPHEIILYYY ELYPTMTVVV VSVATFILLS MVGMAAGMCM CARRRCITPY ELTP GATVP FLLSLICCIR TAKA |
-Macromolecule #3: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 3 / Number of copies: 15 / Enantiomer: LEVO / EC number: togavirin |
|---|---|
| Source (natural) | Organism:   Chikungunya virus Chikungunya virus |
| Molecular weight | Theoretical: 16.428607 KDa |
| Sequence | String: NDCIFEVKHE GKVTGYACLV GDKVMKPAHV KGTIDNADLA KLAFKRSSKY DLECAQIPVH MKSDASKFTH EKPEGYYNWH HGAVQYSGG RFTIPTGAGK PGDSGRPIFD NKGRVVAIVL GGANEGARTA LSVVTWNKDI VTKITPEGAE EW UniProtKB: Structural polyprotein |
-Macromolecule #4: Fab heavy chain
| Macromolecule | Name: Fab heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.87267 KDa |
| Sequence | String: VQLQQSGAEL VKPGASVKIS CKASGYAFSS YWMNWVKQRP GKGLEWIGQI YPGDGDTNYN GKFKGKATLT ADKSSSTAYM QLSSLTSED SAVYFCARGG LTIDYWGQGT TLTVSSAKTT APSVYPLAPV CGGTTGSSVT LGCLVKGYFP EPVTLTWNSG S LSSGVHTF ...String: VQLQQSGAEL VKPGASVKIS CKASGYAFSS YWMNWVKQRP GKGLEWIGQI YPGDGDTNYN GKFKGKATLT ADKSSSTAYM QLSSLTSED SAVYFCARGG LTIDYWGQGT TLTVSSAKTT APSVYPLAPV CGGTTGSSVT LGCLVKGYFP EPVTLTWNSG S LSSGVHTF PALLQSGLYT LSSSVTVTSN TWPSQTITCN VAHPASSTKV DKKIESRR |
-Macromolecule #5: Fab light chain
| Macromolecule | Name: Fab light chain / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.38157 KDa |
| Sequence | String: DIVLTQSPAT LSVTPGDSVS LSCRASQSIS DNLHWYQQKS HESPGLLIKY ASQSISGIPS RFSGSGSGTD FTLSINSVET EDFGMYFCQ QSNSWPYTFG GGTKLEIKRA DAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQNGV L NSWTDQDS ...String: DIVLTQSPAT LSVTPGDSVS LSCRASQSIS DNLHWYQQKS HESPGLLIKY ASQSISGIPS RFSGSGSGTD FTLSINSVET EDFGMYFCQ QSNSWPYTFG GGTKLEIKRA DAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQNGV L NSWTDQDS KDSTYSMSST LTLTKDEYER HNSYTCEATH KTSTSPIVKS FNRNE |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)