[English] 日本語
 Yorodumi
Yorodumi- EMDB-1078: Lumbricus terrestris hemoglobin--the architecture of linker chain... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1078 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Lumbricus terrestris hemoglobin--the architecture of linker chains and structural variation of the central toroid. | |||||||||
 Map data Map data | Whole structure of Lumbricus terrestris hemoglobin | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Lumbricus terrestris (common earthworm) Lumbricus terrestris (common earthworm) | |||||||||
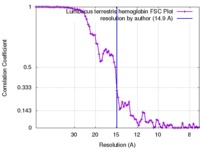
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 14.9 Å | |||||||||
 Authors Authors | Mouche F / Boisset N / Penczek PA | |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2001 Journal: J Struct Biol / Year: 2001Title: Lumbricus terrestris hemoglobin--the architecture of linker chains and structural variation of the central toroid. Authors: F Mouche / N Boisset / P A Penczek /  Abstract: The extracellular giant hemoglobin from the earthworm Lumbricus terrestris was reconstructed at 14.9-A resolution from cryo-electron microscope images, using a new procedure for estimating parameters ...The extracellular giant hemoglobin from the earthworm Lumbricus terrestris was reconstructed at 14.9-A resolution from cryo-electron microscope images, using a new procedure for estimating parameters of the contrast transfer (CTF) function. In this approach, two important CTF parameters, defocus and amplitude contrast ratio, can be refined iteratively within the framework of 3D projection alignment procedure, using minimization of sign disagreement between theoretical CTF and cross-resolution curves. The 3D cryo-EM map is in overall good agreement with the recent X-ray crystallography map of Royer et al. (2000, Proc. Natl. Acad. Sci. USA 97, 7107-7111), and it reveals the local threefold arrangement of the three linker chains present within each 1/12 of the complex. The 144 globin chains and 36 linker chains within the complex are clearly visible, and the interdigitation of the 12 coiled-coil helical spokes forming the central toroidal piece is confirmed. Based on these findings, two mechanisms of the dodecameric unit assembly are proposed and termed "zigzag" and "pairwise" polymerizations. However, the detection by cryo-EM of 12 additional rod-like bodies within the toroid raises the possibility that the architecture of the toroid is more complex than previously thought or that yet unknown ligands or allosteric effectors for this oxygen carrier are present. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1078.map.gz emd_1078.map.gz | 1.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1078-v30.xml emd-1078-v30.xml emd-1078.xml emd-1078.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_1078_fsc.xml emd_1078_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  1078.gif 1078.gif | 83.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1078 http://ftp.pdbj.org/pub/emdb/structures/EMD-1078 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1078 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1078 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1078.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1078.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Whole structure of Lumbricus terrestris hemoglobin | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.76 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Lumbricus terrestris hemoglobin
| Entire | Name: Lumbricus terrestris hemoglobin |
|---|---|
| Components |
|
-Supramolecule #1000: Lumbricus terrestris hemoglobin
| Supramolecule | Name: Lumbricus terrestris hemoglobin / type: sample / ID: 1000 / Oligomeric state: 12 x 12 mer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 3.6 MDa |
-Macromolecule #1: Lumbricus terrestris hemoglobin
| Macromolecule | Name: Lumbricus terrestris hemoglobin / type: protein_or_peptide / ID: 1 / Number of copies: 144 / Oligomeric state: 12 x 12 mer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Lumbricus terrestris (common earthworm) / synonym: earthworm / Tissue: blood Lumbricus terrestris (common earthworm) / synonym: earthworm / Tissue: blood |
| Molecular weight | Experimental: 3.6 MDa |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.2 / Details: 50 mM Tris-HCl, 50 mM MgCl2, 10 mM CaCl2 |
| Staining | Type: NEGATIVE / Details: NO STAIN Cryo-electron microscopy |
| Grid | Details: 400 mesh gold grid |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: home made / Method: Single side blotting and rapid plunging |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2010F |
|---|---|
| Temperature | Min: 86 K / Max: 86 K / Average: 86 K |
| Alignment procedure | Legacy - Electron beam tilt params: 0 |
| Date | Oct 17, 1999 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OPTRONICS / Digitization - Sampling interval: 2.5 µm / Number real images: 16 / Details: Rotating drum microdensitometer / Od range: 1 / Bits/pixel: 16 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 66489 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.0 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.3 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder Specimen holder model: GATAN LIQUID NITROGEN |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)