+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3031 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ATP-bound IstB in complex to duplex DNA | |||||||||
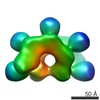
 Map data Map data | IstB decamer bound to duplex DNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ / ATPase / IS21 / DNA transposition / DNA binding / DNA remodeling | |||||||||
| Biological species |   Geobacillus stearothermophilus (bacteria) / synthetic construct (others) Geobacillus stearothermophilus (bacteria) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.5 Å | |||||||||
 Authors Authors | Arias-Palomo E / Berger JM | |||||||||
 Citation Citation |  Journal: Cell / Year: 2015 Journal: Cell / Year: 2015Title: An Atypical AAA+ ATPase Assembly Controls Efficient Transposition through DNA Remodeling and Transposase Recruitment. Authors: Ernesto Arias-Palomo / James M Berger /  Abstract: Transposons are ubiquitous genetic elements that drive genome rearrangements, evolution, and the spread of infectious disease and drug-resistance. Many transposons, such as Mu, Tn7, and IS21, require ...Transposons are ubiquitous genetic elements that drive genome rearrangements, evolution, and the spread of infectious disease and drug-resistance. Many transposons, such as Mu, Tn7, and IS21, require regulatory AAA+ ATPases for function. We use X-ray crystallography and cryo-electron microscopy to show that the ATPase subunit of IS21, IstB, assembles into a clamshell-shaped decamer that sandwiches DNA between two helical pentamers of ATP-associated AAA+ domains, sharply bending the duplex into a 180° U-turn. Biochemical studies corroborate key features of the structure and further show that the IS21 transposase, IstA, recognizes the IstB•DNA complex and promotes its disassembly by stimulating ATP hydrolysis. Collectively, these studies reveal a distinct manner of higher-order assembly and client engagement by a AAA+ ATPase and suggest a mechanistic model where IstB binding and subsequent DNA bending primes a selected insertion site for efficient transposition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3031.map.gz emd_3031.map.gz | 789.6 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3031-v30.xml emd-3031-v30.xml emd-3031.xml emd-3031.xml | 8.7 KB 8.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3031.png emd_3031.png | 557.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3031 http://ftp.pdbj.org/pub/emdb/structures/EMD-3031 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3031 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3031 | HTTPS FTP |
-Validation report
| Summary document |  emd_3031_validation.pdf.gz emd_3031_validation.pdf.gz | 202.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3031_full_validation.pdf.gz emd_3031_full_validation.pdf.gz | 201.6 KB | Display | |
| Data in XML |  emd_3031_validation.xml.gz emd_3031_validation.xml.gz | 5.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3031 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3031 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3031 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3031 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3031.map.gz / Format: CCP4 / Size: 11.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3031.map.gz / Format: CCP4 / Size: 11.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | IstB decamer bound to duplex DNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : ATP-bound IstB decamer bound to a 60 bp duplex DNA
| Entire | Name: ATP-bound IstB decamer bound to a 60 bp duplex DNA |
|---|---|
| Components |
|
-Supramolecule #1000: ATP-bound IstB decamer bound to a 60 bp duplex DNA
| Supramolecule | Name: ATP-bound IstB decamer bound to a 60 bp duplex DNA / type: sample / ID: 1000 / Details: The sample was monodisperse Oligomeric state: One IstB pentamer of dimers binds to a 50-basepair DNA Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 320 KDa / Theoretical: 320 KDa / Method: Analytical size exclusion chromatography |
-Macromolecule #1: IstB
| Macromolecule | Name: IstB / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Oligomeric state: Decamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria) |
| Molecular weight | Theoretical: 29 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #2: Duplex DNA
| Macromolecule | Name: Duplex DNA / type: dna / ID: 2 / Classification: DNA / Structure: DOUBLE HELIX / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: CCTGAATTGT TAAAGCCGAT GATAAAATCC CCAATATAGC CGGAATAAAA TTCCCCACTT |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.012 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20 mM HEPES pH 7.5, 300 mM NaCl, 5 mM MgCl2, 1.5% glycerol, 1 mM DTT, 1 mM ATP |
| Grid | Details: C-Flat 400 mesh 2/2 with thin carbon support |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK II / Method: Blot for 4 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Date | May 23, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 20 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.3 µm / Nominal magnification: 100000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each micrograph |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 8.5 Å / Resolution method: OTHER / Software - Name: EMAN2, SPARX / Number images used: 263191 |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















