+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30189 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The mitochondrial SAM complex from S.cere | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Mitochondria / TRANSLOCASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationmitochondrial outer membrane translocase complex assembly / SAM complex / protein insertion into mitochondrial outer membrane / outer membrane / protein import into mitochondrial matrix / protein transport / mitochondrial outer membrane / mitochondrion Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
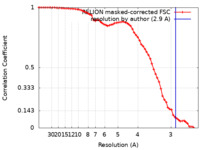
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | ||||||||||||
 Authors Authors | Takeda H / Tsutsumi A | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Mitochondrial sorting and assembly machinery operates by β-barrel switching. Authors: Hironori Takeda / Akihisa Tsutsumi / Tomohiro Nishizawa / Caroline Lindau / Jon V Busto / Lena-Sophie Wenz / Lars Ellenrieder / Kenichiro Imai / Sebastian P Straub / Waltraut Mossmann / Jian ...Authors: Hironori Takeda / Akihisa Tsutsumi / Tomohiro Nishizawa / Caroline Lindau / Jon V Busto / Lena-Sophie Wenz / Lars Ellenrieder / Kenichiro Imai / Sebastian P Straub / Waltraut Mossmann / Jian Qiu / Yu Yamamori / Kentaro Tomii / Junko Suzuki / Takeshi Murata / Satoshi Ogasawara / Osamu Nureki / Thomas Becker / Nikolaus Pfanner / Nils Wiedemann / Masahide Kikkawa / Toshiya Endo /     Abstract: The mitochondrial outer membrane contains so-called β-barrel proteins, which allow communication between the cytosol and the mitochondrial interior. Insertion of β-barrel proteins into the outer ...The mitochondrial outer membrane contains so-called β-barrel proteins, which allow communication between the cytosol and the mitochondrial interior. Insertion of β-barrel proteins into the outer membrane is mediated by the multisubunit mitochondrial sorting and assembly machinery (SAM, also known as TOB). Here we use cryo-electron microscopy to determine the structures of two different forms of the yeast SAM complex at a resolution of 2.8-3.2 Å. The dimeric complex contains two copies of the β-barrel channel protein Sam50-Sam50a and Sam50b-with partially open lateral gates. The peripheral membrane proteins Sam35 and Sam37 cap the Sam50 channels from the cytosolic side, and are crucial for the structural and functional integrity of the dimeric complex. In the second complex, Sam50b is replaced by the β-barrel protein Mdm10. In cooperation with Sam50a, Sam37 recruits and traps Mdm10 by penetrating the interior of its laterally closed β-barrel from the cytosolic side. The substrate-loaded SAM complex contains one each of Sam50, Sam35 and Sam37, but neither Mdm10 nor a second Sam50, suggesting that Mdm10 and Sam50b function as placeholders for a β-barrel substrate released from Sam50a. Our proposed mechanism for dynamic switching of β-barrel subunits and substrate explains how entire precursor proteins can fold in association with the mitochondrial machinery for β-barrel assembly. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30189.map.gz emd_30189.map.gz | 14.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30189-v30.xml emd-30189-v30.xml emd-30189.xml emd-30189.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30189_fsc.xml emd_30189_fsc.xml | 6.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_30189.png emd_30189.png | 66.3 KB | ||
| Filedesc metadata |  emd-30189.cif.gz emd-30189.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30189 http://ftp.pdbj.org/pub/emdb/structures/EMD-30189 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30189 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30189 | HTTPS FTP |
-Related structure data
| Related structure data |  7btwMC  7btxC  7btyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_30189.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30189.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.245 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SAM complex
| Entire | Name: SAM complex |
|---|---|
| Components |
|
-Supramolecule #1: SAM complex
| Supramolecule | Name: SAM complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Mitochondrial outer membrane beta-barrel protein
| Macromolecule | Name: Mitochondrial outer membrane beta-barrel protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.176824 KDa |
| Sequence | String: TFTAKTGTNF GNDNDAEAYL QFEKLIDKKY LKLPTRVNLE ILRGTKIHSS FLFNSYSSLS PQSILNLKVF SQFYNWNTNK GLDIGQRGA RLSLRYEPLF LHKLLHNPHS NESPTLFHEW FLETCWRSTK ICSQGTSAPY MYSGTMLSQA GDQLRTILGH T FVLDKRDH ...String: TFTAKTGTNF GNDNDAEAYL QFEKLIDKKY LKLPTRVNLE ILRGTKIHSS FLFNSYSSLS PQSILNLKVF SQFYNWNTNK GLDIGQRGA RLSLRYEPLF LHKLLHNPHS NESPTLFHEW FLETCWRSTK ICSQGTSAPY MYSGTMLSQA GDQLRTILGH T FVLDKRDH IMCPTKGSML KWSNELSPGK HLKTQLELNS VKSWMNDDFI TFSTTIKTGY LKNLSSQQSL PVHICDKFQS GG PSDIRGF QTFGLGPRDL YDAVGGDAFV SYGLSVFSRL PWKKVEKSNF RLHWFFNGGK LVNHDNTSLG NCIGQLSKEH STS TGIGLV LRHPMARFEL NFTLPITAHE NDLIRKGFQF GLGLAFL UniProtKB: Mitochondrial outer membrane beta-barrel protein |
-Macromolecule #2: Sorting assembly machinery 35 kDa subunit
| Macromolecule | Name: Sorting assembly machinery 35 kDa subunit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.44607 KDa |
| Sequence | String: MVSSFSVPMP VKRIFDTFPL QTYAAQTDKD EAVALEIQRR SYTFTERGGG SSELTVEGTY KLGVYNVFLE ANTGAALATD PWCLFVQLA LCQKNGLVLP THSQEQTPSH TCNHEMLVLS RLSNPDEALP ILVEGYKKRI IRSTVAISEI MRSRILDDAE Q LMYYTLLD ...String: MVSSFSVPMP VKRIFDTFPL QTYAAQTDKD EAVALEIQRR SYTFTERGGG SSELTVEGTY KLGVYNVFLE ANTGAALATD PWCLFVQLA LCQKNGLVLP THSQEQTPSH TCNHEMLVLS RLSNPDEALP ILVEGYKKRI IRSTVAISEI MRSRILDDAE Q LMYYTLLD TVLYDCWITQ IIFCASDAQF MELYSCQKLS GSIVTPLDVE NSLLQKLSAK SLKISLTKRN KFQFRHREIV KS MQGVYHN HHNSVNQEQV LNVLFENSKQ VLLGLKDMLK SDGQPTYLHL KIASYILCIT NVKEPIKLKT FVENECKELV QFA QDTLKN FVQ UniProtKB: Sorting assembly machinery 35 kDa subunit |
-Macromolecule #3: SAM37 isoform 1
| Macromolecule | Name: SAM37 isoform 1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.43759 KDa |
| Sequence | String: MVKGSVHLWG KDGEASLISV DSIALVWFIK LCTSEEAKSM VAGLQIVFSN NTDLSSDGKL PVLILDNGTK VSGYVNIVQF LHKNICTSK YEKGTDYEED LAIVGKKDRL LEYSLLNYVD VEISRLTDYQ LFLNTKNYNE YTKKLFSKLL YFPMWYNTPL Q LRSQAREN ...String: MVKGSVHLWG KDGEASLISV DSIALVWFIK LCTSEEAKSM VAGLQIVFSN NTDLSSDGKL PVLILDNGTK VSGYVNIVQF LHKNICTSK YEKGTDYEED LAIVGKKDRL LEYSLLNYVD VEISRLTDYQ LFLNTKNYNE YTKKLFSKLL YFPMWYNTPL Q LRSQAREN CEEIIGSLTL EDDEEFVESK AMESASQLAQ SKTFKIAHKN KIKGKQELQQ VKYNLQFDNR LQSCVSNWLA AR KKLDDSV ILSSDLLFLA NLYVQLGLPD GNRIRSKLEQ TFGSELLNSM SNKIDDFVHR PSNNLEQRDP QFREQGNVVM SLY NLACKY I UniProtKB: SAM37 isoform 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 299 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)