[English] 日本語
 Yorodumi
Yorodumi- EMDB-2944: Structural characterization of the Olfactomedin-1 disulfide-linke... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2944 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural characterization of the Olfactomedin-1 disulfide-linked tetramer | |||||||||
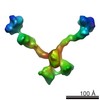
 Map data Map data | Subtomogram of an Olfactomedin-1 tetramer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Olfactomedin-1 (Olfm1) / neurobiology / tetramer / X-ray crystallography / small-angle X-ray scattering (SAXS) / electron tomography / analytical ultracentrifugation (AUC) / coiled coil / calcium / disulfide | |||||||||
| Biological species |  | |||||||||
| Method | electron tomography / negative staining | |||||||||
 Authors Authors | Sharp TH / Pronker MF / Bos TG / Thies-Weesie DM / Janssen BJC | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2015 Journal: J Biol Chem / Year: 2015Title: Olfactomedin-1 Has a V-shaped Disulfide-linked Tetrameric Structure. Authors: Matti F Pronker / Trusanne G A A Bos / Thomas H Sharp / Dominique M E Thies-Weesie / Bert J C Janssen /  Abstract: Olfactomedin-1 (Olfm1; also known as noelin and pancortin) is a member of the olfactomedin domain-containing superfamily and a highly expressed neuronal glycoprotein important for nervous system ...Olfactomedin-1 (Olfm1; also known as noelin and pancortin) is a member of the olfactomedin domain-containing superfamily and a highly expressed neuronal glycoprotein important for nervous system development. It binds a number of secreted proteins and cell surface-bound receptors to induce cell signaling processes. Using a combined approach of x-ray crystallography, solution scattering, analytical ultracentrifugation, and electron microscopy we determined that full-length Olfm1 forms disulfide-linked tetramers with a distinctive V-shaped architecture. The base of the "V" is formed by two disulfide-linked dimeric N-terminal domains. Each of the two V legs consists of a parallel dimeric disulfide-linked coiled coil with a C-terminal β-propeller dimer at the tips. This agrees with our crystal structure of a C-terminal coiled-coil segment and β-propeller combination (Olfm1(coil-Olf)) that reveals a disulfide-linked dimeric arrangement with the β-propeller top faces in an outward exposed orientation. Similar to its family member myocilin, Olfm1 is stabilized by calcium. The dimer-of-dimers architecture suggests a role for Olfm1 in clustering receptors to regulate signaling and sheds light on the conformation of several other olfactomedin domain family members. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2944.map.gz emd_2944.map.gz | 418.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2944-v30.xml emd-2944-v30.xml emd-2944.xml emd-2944.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
| Images |  2944.png 2944.png | 73.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2944 http://ftp.pdbj.org/pub/emdb/structures/EMD-2944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2944 | HTTPS FTP |
-Validation report
| Summary document |  emd_2944_validation.pdf.gz emd_2944_validation.pdf.gz | 126 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2944_full_validation.pdf.gz emd_2944_full_validation.pdf.gz | 125.1 KB | Display | |
| Data in XML |  emd_2944_validation.xml.gz emd_2944_validation.xml.gz | 5.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2944 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2944 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2944 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2944 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2944.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2944.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram of an Olfactomedin-1 tetramer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.57 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Olfactomedin-1 disulfide-linked tetramer
| Entire | Name: Olfactomedin-1 disulfide-linked tetramer |
|---|---|
| Components |
|
-Supramolecule #1000: Olfactomedin-1 disulfide-linked tetramer
| Supramolecule | Name: Olfactomedin-1 disulfide-linked tetramer / type: sample / ID: 1000 / Oligomeric state: Tetramer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 242 KDa / Theoretical: 256 KDa / Method: AUC |
-Supramolecule #1: Olfactomedin-1
| Supramolecule | Name: Olfactomedin-1 / type: organelle_or_cellular_component / ID: 1 / Name.synonym: Olfm1, noelin, pancortin Details: Purified recombinant full-length glycoslated Olfactomedin-1 tetramers were negatively stained with uranyl formate. Number of copies: 1 / Oligomeric state: Tetramer / Recombinant expression: Yes |
|---|---|
| Ref INTERPRO | 0: IPR022082 |
| Ref INTERPRO | 0: IPR003112 |
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 242 KDa / Theoretical: 256 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293ES / Recombinant plasmid: pUPE107.03 Homo sapiens (human) / Recombinant cell: HEK293ES / Recombinant plasmid: pUPE107.03 |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.065 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 150 mM NaCl, 20 mM HEPES |
| Staining | Type: NEGATIVE Details: Full-length Olfm1 was adsorbed to grids for 30 sec. Grids were briefly washed with water and then stained for 30 sec with a freshly prepared filtered 2% uranyl formate solution. |
| Grid | Details: Carbon-coated mesh copper grids glow discharged for 15 sec |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 30,000 times magnification |
| Details | Weak beam illumination |
| Date | Feb 12, 2015 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 58 / Bits/pixel: 32 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 30000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt series - Axis1 - Min angle: -58 ° / Tilt series - Axis1 - Max angle: 58 ° / Tilt series - Axis1 - Angle increment: 2 ° |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Tilt series were collected from -58 to 58 degrees in 2 degree increments. The first and last image was discarded. CTF correction was performed on each projection |
|---|---|
| Final reconstruction | Algorithm: OTHER / Software - Name: IMOD, EMAN2 Details: Sub-tomogram particles were manually picked using e2spt_boxer.py from EMAN2. Each particle was normalized and masked with a sharp spherical mask to remove background density not associated ...Details: Sub-tomogram particles were manually picked using e2spt_boxer.py from EMAN2. Each particle was normalized and masked with a sharp spherical mask to remove background density not associated with the protein. Particles were then filtered to 20 A with a low-pass Gaussian filter, before a tight mask was applied to the remaining density using e2proc3d.py from EMAN2. Number images used: 57 |
| CTF correction | Details: IMOD, each tilt image |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Each coiled-coil dimer of the tetramer was fitted as a rigid body. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)