[English] 日本語
 Yorodumi
Yorodumi- PDB-5amo: Structure of a mouse Olfactomedin-1 disulfide-linked dimer of the... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5amo | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a mouse Olfactomedin-1 disulfide-linked dimer of the Olfactomedin domain and part of the coiled coil | |||||||||
 Components Components | NOELIN | |||||||||
 Keywords Keywords | SIGNALING PROTEIN / OLFM1 / DISULFIDE / NEUROBIOLOGY / DEVELOPMENT / AMPA RECEPTOR / BETA PROPELLER | |||||||||
| Function / homology |  Function and homology information Function and homology informationatrioventricular valve formation / neuronal signal transduction / extrinsic component of postsynaptic density membrane / extrinsic component of synaptic membrane / regulation of amyloid-beta formation / cardiac epithelial to mesenchymal transition / regulation of axon extension / negative regulation of amyloid-beta formation / AMPA glutamate receptor complex / positive regulation of epithelial to mesenchymal transition ...atrioventricular valve formation / neuronal signal transduction / extrinsic component of postsynaptic density membrane / extrinsic component of synaptic membrane / regulation of amyloid-beta formation / cardiac epithelial to mesenchymal transition / regulation of axon extension / negative regulation of amyloid-beta formation / AMPA glutamate receptor complex / positive regulation of epithelial to mesenchymal transition / axonal growth cone / synaptic membrane / amyloid-beta binding / perikaryon / positive regulation of apoptotic process / axon / negative regulation of gene expression / neuronal cell body / positive regulation of gene expression / glutamatergic synapse / endoplasmic reticulum / extracellular space / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.4 Å MOLECULAR REPLACEMENT / Resolution: 2.4 Å | |||||||||
 Authors Authors | Pronker, M.F. / Bos, T.G.A.A. / Sharp, T.H. / Thies-Weesie, D.M. / Janssen, B.J.C. | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2015 Journal: J Biol Chem / Year: 2015Title: Olfactomedin-1 Has a V-shaped Disulfide-linked Tetrameric Structure. Authors: Matti F Pronker / Trusanne G A A Bos / Thomas H Sharp / Dominique M E Thies-Weesie / Bert J C Janssen /  Abstract: Olfactomedin-1 (Olfm1; also known as noelin and pancortin) is a member of the olfactomedin domain-containing superfamily and a highly expressed neuronal glycoprotein important for nervous system ...Olfactomedin-1 (Olfm1; also known as noelin and pancortin) is a member of the olfactomedin domain-containing superfamily and a highly expressed neuronal glycoprotein important for nervous system development. It binds a number of secreted proteins and cell surface-bound receptors to induce cell signaling processes. Using a combined approach of x-ray crystallography, solution scattering, analytical ultracentrifugation, and electron microscopy we determined that full-length Olfm1 forms disulfide-linked tetramers with a distinctive V-shaped architecture. The base of the "V" is formed by two disulfide-linked dimeric N-terminal domains. Each of the two V legs consists of a parallel dimeric disulfide-linked coiled coil with a C-terminal β-propeller dimer at the tips. This agrees with our crystal structure of a C-terminal coiled-coil segment and β-propeller combination (Olfm1(coil-Olf)) that reveals a disulfide-linked dimeric arrangement with the β-propeller top faces in an outward exposed orientation. Similar to its family member myocilin, Olfm1 is stabilized by calcium. The dimer-of-dimers architecture suggests a role for Olfm1 in clustering receptors to regulate signaling and sheds light on the conformation of several other olfactomedin domain family members. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5amo.cif.gz 5amo.cif.gz | 232.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5amo.ent.gz pdb5amo.ent.gz | 184.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5amo.json.gz 5amo.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  5amo_validation.pdf.gz 5amo_validation.pdf.gz | 1.6 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  5amo_full_validation.pdf.gz 5amo_full_validation.pdf.gz | 1.6 MB | Display | |
| Data in XML |  5amo_validation.xml.gz 5amo_validation.xml.gz | 20.5 KB | Display | |
| Data in CIF |  5amo_validation.cif.gz 5amo_validation.cif.gz | 27 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/am/5amo https://data.pdbj.org/pub/pdb/validation_reports/am/5amo ftp://data.pdbj.org/pub/pdb/validation_reports/am/5amo ftp://data.pdbj.org/pub/pdb/validation_reports/am/5amo | HTTPS FTP |
-Related structure data
| Related structure data | 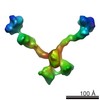 2940C  2941C  2942C  2943C  2944C  4d77S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 54075.621 Da / Num. of mol.: 2 Fragment: COILED COIL AND OLFACTOMEDIN DOMAIN, RESIDUES 17-478 Source method: isolated from a genetically manipulated source Details: N-LINKED GLYCOSYLATION ON RESIDUES ASN473, ASN307 AND ASN394 Source: (gene. exp.)   HOMO SAPIENS (human) / References: UniProt: O88998 HOMO SAPIENS (human) / References: UniProt: O88998 |
|---|
-Sugars , 2 types, 6 molecules 
| #2: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source #3: Sugar | |
|---|
-Non-polymers , 3 types, 30 molecules 




| #4: Chemical | | #5: Chemical | ChemComp-CL / | #6: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|---|
| Sequence details | CORRESPOND |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.9 Å3/Da / Density % sol: 57 % / Description: NONE |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 8.5 Details: SITTING DROP AT 293 K, MIXING PROTEIN AT 6 MG/ML 1:1 WITH PRECIPITANT SOLUTION: 1M LICL, 20% PEG6000 (W/V) AND 100 MM TRIS PH8.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID23-1 / Wavelength: 0.97242 / Beamline: ID23-1 / Wavelength: 0.97242 |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Nov 8, 2013 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97242 Å / Relative weight: 1 |
| Reflection | Resolution: 2.4→95 Å / Num. obs: 26050 / % possible obs: 98.9 % / Observed criterion σ(I): -4 / Redundancy: 3.4 % / Biso Wilson estimate: 49.62 Å2 / Rmerge(I) obs: 0.08 / Net I/σ(I): 7.7 |
| Reflection shell | Resolution: 2.4→2.5 Å / Redundancy: 3.5 % / Rmerge(I) obs: 0.78 / Mean I/σ(I) obs: 1.2 / % possible all: 99.4 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 4D77 Resolution: 2.4→50.318 Å / SU ML: 0.33 / σ(F): 1.92 / Phase error: 32.14 / Stereochemistry target values: ML Details: RESIDUES 17-210, 339-352 AND 478-487 NOT MODELED BECAUSE OF DISORDER OR LIMITED PROTEOLYSIS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 0.4242 Å2 / ksol: 1.3585 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 69.76 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.4→50.318 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj





