[English] 日本語
 Yorodumi
Yorodumi- EMDB-29340: Cryo-EM structure of LRRK2 bound to type I inhibitor LRRK2-IN-1 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of LRRK2 bound to type I inhibitor LRRK2-IN-1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cryo-EM / Parkinson's disease / Kinase / LRRK2 / type I inhibitor / LRRK2-IN-1 / HYDROLASE-HYDROLASE INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationcaveola neck / : / beta-catenin destruction complex binding / regulation of branching morphogenesis of a nerve / Wnt signalosome assembly / regulation of kidney size / regulation of cell projection organization / tangential migration from the subventricular zone to the olfactory bulb / GTP-dependent protein kinase activity / regulation of SNARE complex assembly ...caveola neck / : / beta-catenin destruction complex binding / regulation of branching morphogenesis of a nerve / Wnt signalosome assembly / regulation of kidney size / regulation of cell projection organization / tangential migration from the subventricular zone to the olfactory bulb / GTP-dependent protein kinase activity / regulation of SNARE complex assembly / regulation of neuroblast proliferation / regulation of ER to Golgi vesicle-mediated transport / protein localization to endoplasmic reticulum exit site / peroxidase inhibitor activity / negative regulation of late endosome to lysosome transport / regulation of mitochondrial depolarization / : / positive regulation of dopamine receptor signaling pathway / amphisome / regulation of synaptic vesicle transport / regulation of lysosomal lumen pH / regulation of CAMKK-AMPK signaling cascade / co-receptor binding / negative regulation of GTPase activity / regulation of dopamine receptor signaling pathway / positive regulation of microglial cell activation / regulation of neuron maturation / regulation of retrograde transport, endosome to Golgi / positive regulation of synaptic vesicle endocytosis / cytoplasmic side of mitochondrial outer membrane / negative regulation of autophagosome assembly / olfactory bulb development / JUN kinase kinase kinase activity / regulation of cAMP/PKA signal transduction / neuron projection arborization / striatum development / multivesicular body, internal vesicle / negative regulation of excitatory postsynaptic potential / regulation of dendritic spine morphogenesis / mitochondrion localization / protein localization to mitochondrion / cellular response to dopamine / positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway / endoplasmic reticulum organization / positive regulation of protein autoubiquitination / negative regulation of protein processing / Wnt signalosome / positive regulation of programmed cell death / GTP metabolic process / regulation of canonical Wnt signaling pathway / syntaxin-1 binding / regulation of reactive oxygen species metabolic process / lysosome organization / Golgi-associated vesicle / clathrin binding / PTK6 promotes HIF1A stabilization / negative regulation of macroautophagy / regulation of mitochondrial fission / regulation of locomotion / protein kinase A binding / neuromuscular junction development / regulation of synaptic vesicle exocytosis / intracellular distribution of mitochondria / Golgi organization / microvillus / exploration behavior / endoplasmic reticulum exit site / autolysosome / locomotory exploration behavior / negative regulation of Notch signaling pathway / MAP kinase kinase kinase activity / canonical Wnt signaling pathway / regulation of synaptic vesicle endocytosis / regulation of synaptic transmission, glutamatergic / negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway / Rho protein signal transduction / presynaptic cytosol / neuron projection morphogenesis / phagocytic vesicle / cellular response to manganese ion / JNK cascade / positive regulation of autophagy / dendrite cytoplasm / GTPase activator activity / tubulin binding / cellular response to starvation / positive regulation of protein ubiquitination / SNARE binding / determination of adult lifespan / mitochondrion organization / cellular response to reactive oxygen species / excitatory postsynaptic potential / regulation of membrane potential / trans-Golgi network / calcium-mediated signaling / regulation of protein stability / regulation of autophagy / autophagy / small GTPase binding / mitochondrial membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.52 Å | |||||||||
 Authors Authors | Zhu H / Sun J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2024 Journal: Cell Discov / Year: 2024Title: Pharmacology of LRRK2 with type I and II kinase inhibitors revealed by cryo-EM. Authors: Hanwen Zhu / Patricia Hixson / Wen Ma / Ji Sun /  Abstract: LRRK2 is one of the most promising drug targets for Parkinson's disease. Though type I kinase inhibitors of LRRK2 are under clinical trials, alternative strategies like type II inhibitors are being ...LRRK2 is one of the most promising drug targets for Parkinson's disease. Though type I kinase inhibitors of LRRK2 are under clinical trials, alternative strategies like type II inhibitors are being actively pursued due to the potential undesired effects of type I inhibitors. Currently, a robust method for LRRK2-inhibitor structure determination to guide structure-based drug discovery is lacking, and inhibition mechanisms of available compounds are also unclear. Here we present near-atomic-resolution structures of LRRK2 with type I (LRRK2-IN-1 and GNE-7915) and type II (rebastinib, ponatinib, and GZD-824) inhibitors, uncovering the structural basis of LRRK2 inhibition and conformational plasticity of the kinase domain with molecular dynamics (MD) simulations. Type I and II inhibitors bind to LRRK2 in active-like and inactive conformations, so LRRK2-inhibitor complexes further reveal general structural features associated with LRRK2 activation. Our study provides atomic details of LRRK2-inhibitor interactions and a framework for understanding LRRK2 activation and for rational drug design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29340.map.gz emd_29340.map.gz | 86.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29340-v30.xml emd-29340-v30.xml emd-29340.xml emd-29340.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29340.png emd_29340.png | 35.7 KB | ||
| Filedesc metadata |  emd-29340.cif.gz emd-29340.cif.gz | 6.6 KB | ||
| Others |  emd_29340_half_map_1.map.gz emd_29340_half_map_1.map.gz emd_29340_half_map_2.map.gz emd_29340_half_map_2.map.gz | 84.6 MB 84.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29340 http://ftp.pdbj.org/pub/emdb/structures/EMD-29340 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29340 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29340 | HTTPS FTP |
-Related structure data
| Related structure data |  8fo7MC  8u7hC  8u7lC  8u8aC  8u8bC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29340.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29340.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.324 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29340_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29340_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : LRRK2-LRRK2-IN-1
| Entire | Name: LRRK2-LRRK2-IN-1 |
|---|---|
| Components |
|
-Supramolecule #1: LRRK2-LRRK2-IN-1
| Supramolecule | Name: LRRK2-LRRK2-IN-1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Leucine-rich repeat serine/threonine-protein kinase 2
| Macromolecule | Name: Leucine-rich repeat serine/threonine-protein kinase 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 136.873562 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: KKAVPYNRMK LMIVGNTGSG KTTLLQQLMK TKKSDLGMQS ATVGIDVKDW PIQIRDKRKR DLVLNVWDFA GREEFYSTHP HFMTQRALY LAVYDLSKGQ AEVDAMKPWL FNIKARASSS PVILVGTHLD VSDEKQRKAC MSKITKELLN KRGFPAIRDY H FVNATEES ...String: KKAVPYNRMK LMIVGNTGSG KTTLLQQLMK TKKSDLGMQS ATVGIDVKDW PIQIRDKRKR DLVLNVWDFA GREEFYSTHP HFMTQRALY LAVYDLSKGQ AEVDAMKPWL FNIKARASSS PVILVGTHLD VSDEKQRKAC MSKITKELLN KRGFPAIRDY H FVNATEES DALAKLRKTI INESLNFKIR DQLVVGQLIP DCYVELEKII LSERKNVPIE FPVIDRKRLL QLVRENQLQL DE NELPHAV HFLNESGVLL HFQDPALQLS DLYFVEPKWL CKIMAQILTV KVEGCPKHPK GIISRRDVEK FLSKKRKFPK NYM TQYFKL LEKFQIALPI GEEYLLVPSS LSDHRPVIEL PHCENSEIII RLYEMPYFPM GFWSRLINRL LEISPYMLSG RERA LRPNR RYWRQGIYLN WSPEAYCLVG SEVLDNHPES FLKITVPSCR KGCILLGQVV DHIDSLMEEW FPGLLEIDIC GEGET LLKK WALYSFNDGE EHQKILLDDL MKKAEEGDLL VNPDQPRLTI PISQIAPDLI LADLPRNIML NNDELEFEQA PEFLLG DGS FGSVYRAAYE GEEVAVKIFN KHTSLRLLRQ ELVVLCHLHH PSLISLLAAG IRPRMLVMEL ASKGSLDRLL QQDKASL TR TLQHRIALHV ADGLRYLHSA MIIYRDLKPH NVLLFTLYPN AAIIAKIADY GIAQYCCRMG IKTSEGTPGF RAPEVARG N VIYNQQADVY SFGLLLYDIL TTGGRIVEGL KFPNEFDELE IQGKLPDPVK EYGCAPWPMV EKLIKQCLKE NPQERPTSA QVFDILNSAE LVCLTRRILL PKNVIVECMV ATHHNSRNAS IWLGCGHTDR GQLSFLDLNT EGYTSEEVAD SRILCLALVH LPVEKESWI VSGTQSGTLL VINTEDGKKR HTLEKMTDSV TCLYCNSFSK QSKQKNFLLV GTADGKLAIF EDKTVKLKGA A PLKILNIG NVSTPLMCLS ESTNSTERNV MWGGCGTKIF SFSNDFTIQK LIETRTSQLF SYAAFSDSNI ITVVVDTALY IA KQNSPVV EVWDKKTEKL CGLIDCVHFL REVTVKENKE SKHKMSYSGR VKTLCLQKNT ALWIGTGGGH ILLLDLSTRR LIR VIYNFC NSVRVMMTAQ LGSLKNVMLV LGYNRKNTEG TQKQKEIQSC LTVWDINLPH EVQNLEKHIE VRKELAEKMR RTSV E UniProtKB: Leucine-rich repeat serine/threonine-protein kinase 2 |
-Macromolecule #2: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 1 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Macromolecule #3: 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbon...
| Macromolecule | Name: 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one type: ligand / ID: 3 / Number of copies: 1 / Formula: 4K4 |
|---|---|
| Molecular weight | Theoretical: 570.685 Da |
| Chemical component information | 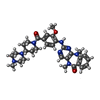 ChemComp-4K4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 62.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)