+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | LRRC8A(T48D):C conformation 2 LRR focus | |||||||||
 Map data Map data | LRRC8A(T48D):C conformation 2 LRR focus | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ION CHANNEL / VOLUME-REGULATION / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationMiscellaneous transport and binding events / pre-B cell differentiation / aspartate transmembrane transport / volume-sensitive anion channel activity / cyclic-GMP-AMP transmembrane transporter activity / cyclic-GMP-AMP transmembrane import across plasma membrane / taurine transmembrane transport / monoatomic anion transmembrane transport / protein hexamerization / cellular response to osmotic stress ...Miscellaneous transport and binding events / pre-B cell differentiation / aspartate transmembrane transport / volume-sensitive anion channel activity / cyclic-GMP-AMP transmembrane transporter activity / cyclic-GMP-AMP transmembrane import across plasma membrane / taurine transmembrane transport / monoatomic anion transmembrane transport / protein hexamerization / cellular response to osmotic stress / monoatomic anion transport / cell volume homeostasis / response to osmotic stress / intracellular glucose homeostasis / monoatomic ion channel complex / positive regulation of myoblast differentiation / fat cell differentiation / chloride transmembrane transport / positive regulation of insulin secretion / spermatogenesis / lysosomal membrane / endoplasmic reticulum membrane / cell surface / endoplasmic reticulum / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Kern DM / Brohawn SG | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels. Authors: David M Kern / Julia Bleier / Somnath Mukherjee / Jennifer M Hill / Anthony A Kossiakoff / Ehud Y Isacoff / Stephen G Brohawn /  Abstract: Leucine-rich repeat-containing protein 8 (LRRC8) family members form volume-regulated anion channels activated by hypoosmotic cell swelling. LRRC8 channels are ubiquitously expressed in vertebrate ...Leucine-rich repeat-containing protein 8 (LRRC8) family members form volume-regulated anion channels activated by hypoosmotic cell swelling. LRRC8 channels are ubiquitously expressed in vertebrate cells as heteromeric assemblies of LRRC8A (SWELL1) and LRRC8B-E subunits. Channels of different subunit composition have distinct properties that explain the functional diversity of LRRC8 currents across cell types. However, the basis for heteromeric LRRC8 channel assembly and function is unknown. Here we leverage a fiducial-tagging strategy to determine single-particle cryo-EM structures of heterohexameric LRRC8A:C channels in multiple conformations. Compared to homomers, LRRC8A:C channels show pronounced differences in architecture due to heterotypic LRR interactions that displace subunits away from the conduction axis and poise the channel for activation. Structures and functional studies further reveal that lipids embedded in the channel pore block ion conduction in the closed state. These results provide insight into determinants for heteromeric LRRC8 channel assembly, activity and gating by lipids. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28895.map.gz emd_28895.map.gz | 259.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28895-v30.xml emd-28895-v30.xml emd-28895.xml emd-28895.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28895.png emd_28895.png | 75 KB | ||
| Filedesc metadata |  emd-28895.cif.gz emd-28895.cif.gz | 6.2 KB | ||
| Others |  emd_28895_additional_1.map.gz emd_28895_additional_1.map.gz emd_28895_half_map_1.map.gz emd_28895_half_map_1.map.gz emd_28895_half_map_2.map.gz emd_28895_half_map_2.map.gz | 137.5 MB 255.1 MB 255.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28895 http://ftp.pdbj.org/pub/emdb/structures/EMD-28895 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28895 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28895 | HTTPS FTP |
-Related structure data
| Related structure data |  8f75MC  8dr8C  8draC  8dreC  8drkC  8drnC  8droC  8drqC  8ds3C  8ds9C  8dsaC  8f74C  8f77C  8f79C  8f7bC  8f7dC  8f7eC  8f7jC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28895.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28895.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LRRC8A(T48D):C conformation 2 LRR focus | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.048 Å | ||||||||||||||||||||||||||||||||||||
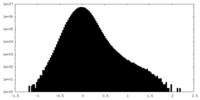
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: LRRC8A(T48D):C conformation 2 LRR focus unsharpened
| File | emd_28895_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LRRC8A(T48D):C conformation 2 LRR focus unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
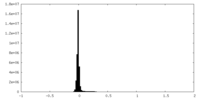
| Density Histograms |
-Half map: LRRC8A(T48D):C conformation 2 LRR focus
| File | emd_28895_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LRRC8A(T48D):C conformation 2 LRR focus | ||||||||||||
| Projections & Slices |
| ||||||||||||
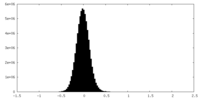
| Density Histograms |
-Half map: LRRC8A(T48D):C conformation 2 LRR focus
| File | emd_28895_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LRRC8A(T48D):C conformation 2 LRR focus | ||||||||||||
| Projections & Slices |
| ||||||||||||
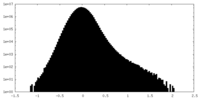
| Density Histograms |
- Sample components
Sample components
-Entire : LRRC8A:C
| Entire | Name: LRRC8A:C |
|---|---|
| Components |
|
-Supramolecule #1: LRRC8A:C
| Supramolecule | Name: LRRC8A:C / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 620 KDa |
-Macromolecule #1: Volume-regulated anion channel subunit LRRC8A
| Macromolecule | Name: Volume-regulated anion channel subunit LRRC8A / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 85.002633 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DTGPTGIKYD LDRHQYNYVD AVCYENRLHW FAKYFPYLVL LHTLIFLACS NFWFKFPRTS SKLEHFVSIL LKCFDSPWTT RALSETVVE ESDPKPAFSK MNGSMDKKSS TVSEDVEATV PMLQRTKSRI EQGIVDRSET GVLDKKEGEQ AKALFEKVKK F RTHVEEGD ...String: DTGPTGIKYD LDRHQYNYVD AVCYENRLHW FAKYFPYLVL LHTLIFLACS NFWFKFPRTS SKLEHFVSIL LKCFDSPWTT RALSETVVE ESDPKPAFSK MNGSMDKKSS TVSEDVEATV PMLQRTKSRI EQGIVDRSET GVLDKKEGEQ AKALFEKVKK F RTHVEEGD IVYRLYMRQT IIKVIKFALI ICYTVYYVHN IKFDVDCTVD IESLTGYRTY RCAHPLATLF KILASFYISL VI FYGLICM YTLWWMLRRS LKKYSFESIR EESSYSDIPD VKNDFAFMLH LIDQYDPLYS KRFAVFLSEV SENKLRQLNL NNE WTLDKL RQRLTKNAQD KLELHLFMLS GIPDTVFDLV ELEVLKLELI PDVTIPPSIA QLTGLKELWL YHTAAKIEAP ALAF LRENL RALHIKFTDI KEIPLWIYSL KTLEELHLTG NLSAENNRYI VIDGLRELKR LKVLRLKSNL SKLPQVVTDV GVHLQ KLSI NNEGTKLIVL NSLKKMVNLT ELELIRCDLE RIPHSIFSLH NLQEIDLKDN NLKTIEEIIS FQHLHRLTCL KLWYNH IAY IPIQIGNLTN LERLYLNRNK IEKIPTQLFY CRKLRYLDLS HNNLTFLPAD IGLLQNLQNL AVTANRIEAL PPELFQC RK LRALHLGNNV LQSLPSRVGE LTNLTQIELR GNRLECLPVE LGECPLLKRS GLVVEEDLFS TLPPEVKERL WRADKEQA S NSLEVLFQ UniProtKB: Volume-regulated anion channel subunit LRRC8A |
-Macromolecule #2: Volume-regulated anion channel subunit LRRC8C
| Macromolecule | Name: Volume-regulated anion channel subunit LRRC8C / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 93.624594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIPVTEFRQF SEQQPAFRVL KPWWDVFTDY LSVAMLMIGV FGCTLQVMQD KIICLPKRVQ PAQNHSSVPN VSQAVISTTP LPPPKPSPT NPATVEMKGL KTDLDLQQYS FINQMCYERA LHWYAKYFPY LVLIHTLVFM LCSNFWFKFP GSSSKIEHFI S ILGKCFDS ...String: MIPVTEFRQF SEQQPAFRVL KPWWDVFTDY LSVAMLMIGV FGCTLQVMQD KIICLPKRVQ PAQNHSSVPN VSQAVISTTP LPPPKPSPT NPATVEMKGL KTDLDLQQYS FINQMCYERA LHWYAKYFPY LVLIHTLVFM LCSNFWFKFP GSSSKIEHFI S ILGKCFDS PWTTRALSEV SGEDSEEKDN RKNNMNRSGT IQSGPEGNLV RSQSLKSIPE KFVVDKSAAG ALDKKEGEQA KA LFEKVKK FRLHVEEGDI LYAMYVRQTV LKVIKFLIII AYNSALVSKV QFTVDCNVDI QDMTGYKNFS CNHTMAHLFS KLS FCYLCF VSIYGLTCLY TLYWLFYRSL REYSFEYVRQ ETGIDDIPDV KNDFAFMLHM IDQYDPLYSK RFAVFLSEVS ENKL KQLNL NNEWTPDKLR QKLQTNAHNR LELPLIMLSG LPDTVFEITE LQSLKLEIIK NVMIPATIAQ LDNLQELCLH QCSVK IHSA ALSFLKENLK VLSVKFDDMR ELPPWMYGLR NLEELYLVGS LSHDISKNVT LESLRDLKSL KILSIKSNVS KIPQAV VDV SSHLQKMCVH NDGTKLVMLN NLKKMTNLTE LELVHCDLER IPHAVFSLLS LQELDLKENN LKSIEEIVSF QHLRKLT VL KLWYNSIAYI PEHIKKLTSL ERLFFSHNKV EVLPSHLFLC NKIRYLDLSY NDIRFIPPEI GVLQSLQYFS ITCNKVES L PDELYFCKKL KTLKIGKNSL SVLSPKIGNL LFLSYLDIKG NHFEVLPPEL GDCRALKRAG LVVEDALFET LPSDVREQM KADSNSENLY FQG UniProtKB: Volume-regulated anion channel subunit LRRC8C |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 0.002 µm / Nominal defocus min: 0.0006 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 71198 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)