+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Brain Aldehyde Dehydrogenase 1 family, member A1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | human brain / ALDH1A1 / Oxidoreductase | |||||||||
| Function / homology |  Function and homology information Function and homology informationoxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor / aldehyde dehydrogenase (NAD+) / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Tringides ML | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Life Sci Alliance / Year: 2023 Journal: Life Sci Alliance / Year: 2023Title: A cryo-electron microscopic approach to elucidate protein structures from human brain microsomes. Authors: Marios L Tringides / Zhemin Zhang / Christopher E Morgan / Chih-Chia Su / Edward W Yu /  Abstract: We recently developed a "Build and Retrieve" cryo-electron microscopy (cryo-EM) methodology, which is capable of simultaneously producing near-atomic resolution cryo-EM maps for several individual ...We recently developed a "Build and Retrieve" cryo-electron microscopy (cryo-EM) methodology, which is capable of simultaneously producing near-atomic resolution cryo-EM maps for several individual proteins from a heterogeneous, multiprotein sample. Here we report the use of "Build and Retrieve" to define the composition of a raw human brain microsomal lysate. From this sample, we simultaneously identify and solve cryo-EM structures of five different brain enzymes whose functions affect neurotransmitter recycling, iron metabolism, glycolysis, axonal development, energy homeostasis, and retinoic acid biosynthesis. Interestingly, malfunction of these important proteins has been directly linked to several neurodegenerative disorders, such as Alzheimer's, Huntington's, and Parkinson's diseases. Our work underscores the importance of cryo-EM in facilitating tissue and organ proteomics at the atomic level. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27575.map.gz emd_27575.map.gz | 5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27575-v30.xml emd-27575-v30.xml emd-27575.xml emd-27575.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27575.png emd_27575.png | 650.9 KB | ||
| Filedesc metadata |  emd-27575.cif.gz emd-27575.cif.gz | 5.6 KB | ||
| Others |  emd_27575_half_map_1.map.gz emd_27575_half_map_1.map.gz emd_27575_half_map_2.map.gz emd_27575_half_map_2.map.gz | 59.1 MB 59.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27575 http://ftp.pdbj.org/pub/emdb/structures/EMD-27575 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27575 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27575 | HTTPS FTP |
-Related structure data
| Related structure data |  8dnoMC  8dnmC  8dnpC  8dnsC  8dnuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27575.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27575.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
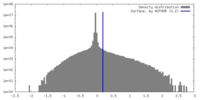
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_27575_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
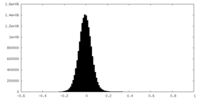
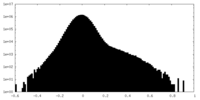
| Density Histograms |
-Half map: #1
| File | emd_27575_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
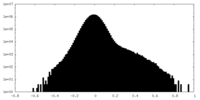
| Density Histograms |
- Sample components
Sample components
-Entire : Aldehyde Dehydrogenase 1 family member A1
| Entire | Name: Aldehyde Dehydrogenase 1 family member A1 |
|---|---|
| Components |
|
-Supramolecule #1: Aldehyde Dehydrogenase 1 family member A1
| Supramolecule | Name: Aldehyde Dehydrogenase 1 family member A1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Retinal dehydrogenase 1
| Macromolecule | Name: Retinal dehydrogenase 1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54.89759 KDa |
| Sequence | String: MSSSGTPDLP VLLTDLKIQY TKIFINNEWH DSVSGKKFPV FNPATEEELC QVEEGDKEDV DKAVKAARQA FQIGSPWRTM DASERGRLL YKLADLIERD RLLLATMESM NGGKLYSNAY LSDLAGCIKT LRYCAGWADK IQGRTIPIDG NFFTYTRHEP I GVCGQIIP ...String: MSSSGTPDLP VLLTDLKIQY TKIFINNEWH DSVSGKKFPV FNPATEEELC QVEEGDKEDV DKAVKAARQA FQIGSPWRTM DASERGRLL YKLADLIERD RLLLATMESM NGGKLYSNAY LSDLAGCIKT LRYCAGWADK IQGRTIPIDG NFFTYTRHEP I GVCGQIIP WNFPLVMLIW KIGPALSCGN TVVVKPAEQT PLTALHVASL IKEAGFPPGV VNIVPGYGPT AGAAISSHMD ID KVAFTGS TEVGKLIKEA AGKSNLKRVT LELGGKSPCI VLADADLDNA VEFAHHGVFY HQGQCCIAAS RIFVEESIYD EFV RRSVER AKKYILGNPL TPGVTQGPQI DKEQYDKILD LIESGKKEGA KLECGGGPWG NKGYFVQPTV FSNVTDEMRI AKEE IFGPV QQIMKFKSLD DVIKRANNTF YGLSAGVFTK DIDKAITISS ALQAGTVWVN CYGVVSAQCP FGGFKMSGNG RELGE YGFH EYTEVKTVTV KISQKNS UniProtKB: aldehyde dehydrogenase (NAD(+)) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | tissue |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 36.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.706 µm / Nominal defocus min: 1.212 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)