[English] 日本語
 Yorodumi
Yorodumi- EMDB-27060: Cryo-EM structure of the supercoiled E. coli K12 flagellar filame... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the supercoiled E. coli K12 flagellar filament core, Normal waveform | |||||||||
 Map data Map data | Cryo-EM structure of the supercoiled E. coli K12 flagellar filament core, Normal waveform | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bacterial motility / flagellar filament / flagellin / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationToll Like Receptor 5 (TLR5) Cascade / MyD88 deficiency (TLR5) / IRAK4 deficiency (TLR5) / MyD88 cascade initiated on plasma membrane / bacterial-type flagellum / structural molecule activity / extracellular region Similarity search - Function | |||||||||
| Biological species |  | |||||||||
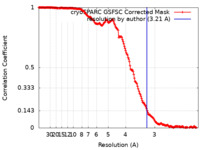
| Method | single particle reconstruction / cryo EM / Resolution: 3.21 Å | |||||||||
 Authors Authors | Sonani RR / Kreutzberger MAB / Sebastian AL / Scharf B / Egelman EH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Convergent evolution in the supercoiling of prokaryotic flagellar filaments. Authors: Mark A B Kreutzberger / Ravi R Sonani / Junfeng Liu / Sharanya Chatterjee / Fengbin Wang / Amanda L Sebastian / Priyanka Biswas / Cheryl Ewing / Weili Zheng / Frédéric Poly / Gad Frankel / ...Authors: Mark A B Kreutzberger / Ravi R Sonani / Junfeng Liu / Sharanya Chatterjee / Fengbin Wang / Amanda L Sebastian / Priyanka Biswas / Cheryl Ewing / Weili Zheng / Frédéric Poly / Gad Frankel / B F Luisi / Chris R Calladine / Mart Krupovic / Birgit E Scharf / Edward H Egelman /    Abstract: The supercoiling of bacterial and archaeal flagellar filaments is required for motility. Archaeal flagellar filaments have no homology to their bacterial counterparts and are instead homologs of ...The supercoiling of bacterial and archaeal flagellar filaments is required for motility. Archaeal flagellar filaments have no homology to their bacterial counterparts and are instead homologs of bacterial type IV pili. How these prokaryotic flagellar filaments, each composed of thousands of copies of identical subunits, can form stable supercoils under torsional stress is a fascinating puzzle for which structural insights have been elusive. Advances in cryoelectron microscopy (cryo-EM) make it now possible to directly visualize the basis for supercoiling, and here, we show the atomic structures of supercoiled bacterial and archaeal flagellar filaments. For the bacterial flagellar filament, we identify 11 distinct protofilament conformations with three broad classes of inter-protomer interface. For the archaeal flagellar filament, 10 protofilaments form a supercoil geometry supported by 10 distinct conformations, with one inter-protomer discontinuity creating a seam inside of the curve. Our results suggest that convergent evolution has yielded stable superhelical geometries that enable microbial locomotion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27060.map.gz emd_27060.map.gz | 481.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27060-v30.xml emd-27060-v30.xml emd-27060.xml emd-27060.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27060_fsc.xml emd_27060_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_27060.png emd_27060.png | 110.1 KB | ||
| Filedesc metadata |  emd-27060.cif.gz emd-27060.cif.gz | 5 KB | ||
| Others |  emd_27060_half_map_1.map.gz emd_27060_half_map_1.map.gz emd_27060_half_map_2.map.gz emd_27060_half_map_2.map.gz | 474.4 MB 474.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27060 http://ftp.pdbj.org/pub/emdb/structures/EMD-27060 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27060 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27060 | HTTPS FTP |
-Related structure data
| Related structure data |  8cxmMC  8cviC  8cwmC  8cyeC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27060.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27060.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the supercoiled E. coli K12 flagellar filament core, Normal waveform | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM structure of the supercoiled E. coli K12...
| File | emd_27060_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the supercoiled E. coli K12 flagellar filament core, Normal waveform | ||||||||||||
| Projections & Slices |
| ||||||||||||
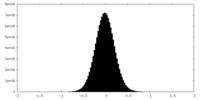
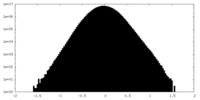
| Density Histograms |
-Half map: Cryo-EM structure of the supercoiled E. coli K12...
| File | emd_27060_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the supercoiled E. coli K12 flagellar filament core, Normal waveform | ||||||||||||
| Projections & Slices |
| ||||||||||||
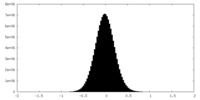
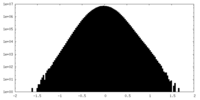
| Density Histograms |
- Sample components
Sample components
-Entire : E. coli K12 flagellar filament core
| Entire | Name: E. coli K12 flagellar filament core |
|---|---|
| Components |
|
-Supramolecule #1: E. coli K12 flagellar filament core
| Supramolecule | Name: E. coli K12 flagellar filament core / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Flagellin
| Macromolecule | Name: Flagellin / type: protein_or_peptide / ID: 1 / Number of copies: 55 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 51.330582 KDa |
| Sequence | String: MAQVINTNSL SLITQNNINK NQSALSSSIE RLSSGLRINS AKDDAAGQAI ANRFTSNIKG LTQAARNAND GISVAQTTEG ALSEINNNL QRVRELTVQA TTGTNSESDL SSIQDEIKSR LDEIDRVSGQ TQFNGVNVLA KNGSMKIQVG ANDNQTITID L KQIDAKTL ...String: MAQVINTNSL SLITQNNINK NQSALSSSIE RLSSGLRINS AKDDAAGQAI ANRFTSNIKG LTQAARNAND GISVAQTTEG ALSEINNNL QRVRELTVQA TTGTNSESDL SSIQDEIKSR LDEIDRVSGQ TQFNGVNVLA KNGSMKIQVG ANDNQTITID L KQIDAKTL GLDGFSVKNN DTVTTSAPVT AFGATTTNNI KLTGITLSTE AATDTGGTNP ASIEGVYTDN GNDYYAKITG GD NDGKYYA VTVANDGTVT MATGATANAT VTDANTTKAT TITSGGTPVQ IDNTAGSATA NLGAVSLVKL QDSKGNDTDT YAL KDTNGN LYAADVNETT GAVSVKTITY TDSSGAASSP TAVKLGGDDG KTEVVDIDGK TYDSADLNGG NLQTGLTAGG EALT AVANG KTTDPLKALD DAIASVDKFR SSLGAVQNRL DSAVTNLNNT TTNLSEAQSR IQDADYATEV SNMSKAQIIQ QAGNS VLAK ANQVPQQVLS LLQG UniProtKB: Flagellin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)