+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Atomic model of the partial VSV nucleocapsid | |||||||||
 Map data Map data | CryoEM density map of partial VSV nucleocapsid. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Vesicular stomatitis virus / Nucleocapsid / Matrix protein M / Nucleocapsid protein N / VIRAL PROTEIN-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA replication / host cell nuclear membrane / helical viral capsid / viral budding via host ESCRT complex / viral transcription / viral nucleocapsid / structural constituent of virion / host cell cytoplasm / ribonucleoprotein complex / viral envelope / RNA binding Similarity search - Function | |||||||||
| Biological species |  Vesicular stomatitis virus Vesicular stomatitis virus | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.47 Å | |||||||||
 Authors Authors | Zhou K / Si Z / Ge P / Tsao J / Luo M / Zhou ZH | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Atomic model of vesicular stomatitis virus and mechanism of assembly. Authors: Kang Zhou / Zhu Si / Peng Ge / Jun Tsao / Ming Luo / Z Hong Zhou /   Abstract: Like other negative-strand RNA viruses (NSVs) such as influenza and rabies, vesicular stomatitis virus (VSV) has a three-layered organization: a layer of matrix protein (M) resides between the ...Like other negative-strand RNA viruses (NSVs) such as influenza and rabies, vesicular stomatitis virus (VSV) has a three-layered organization: a layer of matrix protein (M) resides between the glycoprotein (G)-studded membrane envelope and the nucleocapsid, which is composed of the nucleocapsid protein (N) and the encapsidated genomic RNA. Lack of in situ atomic structures of these viral components has limited mechanistic understanding of assembling the bullet-shaped virion. Here, by cryoEM and sub-particle reconstruction, we have determined the in situ structures of M and N inside VSV at 3.47 Å resolution. In the virion, N and M sites have a stoichiometry of 1:2. The in situ structures of both N and M differ from their crystal structures in their N-terminal segments and oligomerization loops. N-RNA, N-N, and N-M-M interactions govern the formation of the capsid. A double layer of M contributes to packaging of the helical nucleocapsid: the inner M (IM) joins neighboring turns of the N helix, while the outer M (OM) contacts G and the membrane envelope. The pseudo-crystalline organization of G is further mapped by cryoET. The mechanism of VSV assembly is delineated by the network interactions of these viral components. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26841.map.gz emd_26841.map.gz | 14.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26841-v30.xml emd-26841-v30.xml emd-26841.xml emd-26841.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26841.png emd_26841.png | 159.8 KB | ||
| Filedesc metadata |  emd-26841.cif.gz emd-26841.cif.gz | 6.4 KB | ||
| Others |  emd_26841_half_map_1.map.gz emd_26841_half_map_1.map.gz emd_26841_half_map_2.map.gz emd_26841_half_map_2.map.gz | 11.8 MB 11.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26841 http://ftp.pdbj.org/pub/emdb/structures/EMD-26841 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26841 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26841 | HTTPS FTP |
-Validation report
| Summary document |  emd_26841_validation.pdf.gz emd_26841_validation.pdf.gz | 895 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26841_full_validation.pdf.gz emd_26841_full_validation.pdf.gz | 894.5 KB | Display | |
| Data in XML |  emd_26841_validation.xml.gz emd_26841_validation.xml.gz | 9.5 KB | Display | |
| Data in CIF |  emd_26841_validation.cif.gz emd_26841_validation.cif.gz | 10.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26841 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26841 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26841 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26841 | HTTPS FTP |
-Related structure data
| Related structure data |  7uwsMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26841.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26841.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM density map of partial VSV nucleocapsid. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.325 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_26841_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
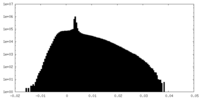
| Projections & Slices |
| ||||||||||||
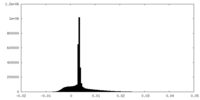
| Density Histograms |
-Half map: #1
| File | emd_26841_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
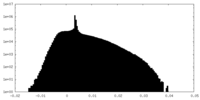
| Projections & Slices |
| ||||||||||||
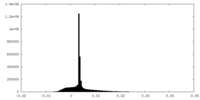
| Density Histograms |
- Sample components
Sample components
-Entire : Vesicular stomatitis virus
| Entire | Name:  Vesicular stomatitis virus Vesicular stomatitis virus |
|---|---|
| Components |
|
-Supramolecule #1: Vesicular stomatitis virus
| Supramolecule | Name: Vesicular stomatitis virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 / NCBI-ID: 11276 / Sci species name: Vesicular stomatitis virus / Sci species strain: San Juan / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 600 KDa |
| Virus shell | Shell ID: 1 / Name: Helical nucleocapsid / Diameter: 700.0 Å |
-Macromolecule #1: Nucleoprotein
| Macromolecule | Name: Nucleoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vesicular stomatitis virus / Strain: San Juan Vesicular stomatitis virus / Strain: San Juan |
| Molecular weight | Theoretical: 47.463949 KDa |
| Sequence | String: MSVTVKRIID NTVIVPKLPA NEDPVEYPAD YFRKSKEIPL YINTTKSLSD LRGYVYQGLK SGNVSIIHVN SYLYGALKDI RGKLDKDWS SFGINIGKAG DTIGIFDLVS LKALDGVLPD GVSDASRTSA DDKWLPLYLL GLYRVGRTQM PEYRKKLMDG L TNQCKMIN ...String: MSVTVKRIID NTVIVPKLPA NEDPVEYPAD YFRKSKEIPL YINTTKSLSD LRGYVYQGLK SGNVSIIHVN SYLYGALKDI RGKLDKDWS SFGINIGKAG DTIGIFDLVS LKALDGVLPD GVSDASRTSA DDKWLPLYLL GLYRVGRTQM PEYRKKLMDG L TNQCKMIN EQFEPLVPEG RDIFDVWGND SNYTKIVAAV DMFFHMFKKH ECASFRYGTI VSRFKDCAAL ATFGHLCKIT GM STEDVTT WILNREVADE MVQMMLPGQE IDKADSYMPY LIDFGLSSKS PYSSVKNPAF HFWGQLTALL LRSTRARNAR QPD DIEYTS LTTAGLLYAY AVGSSADLAQ QFCVGDNKYT PDDSTGGLTT NAPPQGRDVV EWLGWFEDQN RKPTPDMMQY AKRA VMSLQ GLREKTIGKY AKSEFDK UniProtKB: Nucleoprotein |
-Macromolecule #3: Matrix protein
| Macromolecule | Name: Matrix protein / type: protein_or_peptide / ID: 3 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vesicular stomatitis virus Vesicular stomatitis virus |
| Molecular weight | Theoretical: 26.228188 KDa |
| Sequence | String: MSSFKKILGL SSKSHKKSKK LGLPPPYDES SPMEIQPSAP LSNDFFGMED MDLYDKDSLR YEKFRFMLKM TVRSNKPFRS YDDVTAAVS QWDNSYIGMV GKRPFYKIIA LIGSSHLQAT PAVLADLNQP EYYATLTGRC FLPHRLGLIP PMFNVSETFR K PFNIGIYK ...String: MSSFKKILGL SSKSHKKSKK LGLPPPYDES SPMEIQPSAP LSNDFFGMED MDLYDKDSLR YEKFRFMLKM TVRSNKPFRS YDDVTAAVS QWDNSYIGMV GKRPFYKIIA LIGSSHLQAT PAVLADLNQP EYYATLTGRC FLPHRLGLIP PMFNVSETFR K PFNIGIYK GTLDFTFTVS DDESNEKVPH VWEYMNPKYQ SQIQKEGLKF GLILSKKATG TWVLDQLSPF K UniProtKB: Matrix protein |
-Macromolecule #2: RNA
| Macromolecule | Name: RNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Vesicular stomatitis virus / Strain: San Juan Vesicular stomatitis virus / Strain: San Juan |
| Molecular weight | Theoretical: 116.603883 KDa |
| Sequence | String: UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU U UUUUUUUU ...String: UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU U UUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UU UUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUU UUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: PBS buffer |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 2-44 / Number grids imaged: 1 / Number real images: 2008 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 75.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 1.42 Å Applied symmetry - Helical parameters - Δ&Phi: -10.14 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 3.47 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION Details: Additional sub-particle reconstruction was carried out after class3D of helices. Number images used: 23207 |
|---|---|
| Segment selection | Number selected: 7732 |
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)