[English] 日本語
 Yorodumi
Yorodumi- EMDB-26759: Locally refined cryoEM map of Azotobacter vinelandii FeP in a 1:1... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Locally refined cryoEM map of Azotobacter vinelandii FeP in a 1:1 complex (ATP-bound) with MoFeP during catalytic N2 reduction | |||||||||||||||
 Map data Map data | deepEMhancer sharpened map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | nitrogenase / FeP / nitrogen fixation / nitrogenase complex / OXIDOREDUCTASE | |||||||||||||||
| Biological species |  Azotobacter vinelandii DJ (bacteria) Azotobacter vinelandii DJ (bacteria) | |||||||||||||||
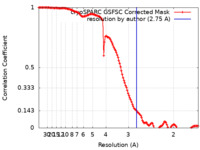
| Method | single particle reconstruction / cryo EM / Resolution: 2.75 Å | |||||||||||||||
 Authors Authors | Rutledge HL / Cook B / Tezcan FA / Herzik MA | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Structures of the nitrogenase complex prepared under catalytic turnover conditions. Authors: Hannah L Rutledge / Brian D Cook / Hoang P M Nguyen / Mark A Herzik / F Akif Tezcan /  Abstract: The enzyme nitrogenase couples adenosine triphosphate (ATP) hydrolysis to the multielectron reduction of atmospheric dinitrogen into ammonia. Despite extensive research, the mechanistic details of ...The enzyme nitrogenase couples adenosine triphosphate (ATP) hydrolysis to the multielectron reduction of atmospheric dinitrogen into ammonia. Despite extensive research, the mechanistic details of ATP-dependent energy transduction and dinitrogen reduction by nitrogenase are not well understood, requiring new strategies to monitor its structural dynamics during catalytic action. Here, we report cryo-electron microscopy structures of the nitrogenase complex prepared under enzymatic turnover conditions. We observe that asymmetry governs all aspects of the nitrogenase mechanism, including ATP hydrolysis, protein-protein interactions, and catalysis. Conformational changes near the catalytic iron-molybdenum cofactor are correlated with the nucleotide-hydrolysis state of the enzyme. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26759.map.gz emd_26759.map.gz | 180 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26759-v30.xml emd-26759-v30.xml emd-26759.xml emd-26759.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26759_fsc.xml emd_26759_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_26759.png emd_26759.png | 55.1 KB | ||
| Masks |  emd_26759_msk_1.map emd_26759_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26759.cif.gz emd-26759.cif.gz | 5.5 KB | ||
| Others |  emd_26759_additional_1.map.gz emd_26759_additional_1.map.gz emd_26759_half_map_1.map.gz emd_26759_half_map_1.map.gz emd_26759_half_map_2.map.gz emd_26759_half_map_2.map.gz | 108.2 MB 200.2 MB 200.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26759 http://ftp.pdbj.org/pub/emdb/structures/EMD-26759 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26759 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26759 | HTTPS FTP |
-Validation report
| Summary document |  emd_26759_validation.pdf.gz emd_26759_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26759_full_validation.pdf.gz emd_26759_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_26759_validation.xml.gz emd_26759_validation.xml.gz | 21.6 KB | Display | |
| Data in CIF |  emd_26759_validation.cif.gz emd_26759_validation.cif.gz | 28 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26759 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26759 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26759 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26759 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26759.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26759.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | deepEMhancer sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26759_msk_1.map emd_26759_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: unsharpened map
| File | emd_26759_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: gold-standard half map A
| File | emd_26759_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | gold-standard half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: gold-standard half map B
| File | emd_26759_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | gold-standard half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Locally refined cryoEM map of Azotobacter vinelandii FeP in a 1:1...
| Entire | Name: Locally refined cryoEM map of Azotobacter vinelandii FeP in a 1:1 complex (ATP-bound) with MoFeP during catalytic N2 reduction |
|---|---|
| Components |
|
-Supramolecule #1: Locally refined cryoEM map of Azotobacter vinelandii FeP in a 1:1...
| Supramolecule | Name: Locally refined cryoEM map of Azotobacter vinelandii FeP in a 1:1 complex (ATP-bound) with MoFeP during catalytic N2 reduction type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Wild-type MoFeP and FeP were purified from the native organism, Azotobacter vinelandii. This map is the locally refined map for the FeP portion of the 1:1 complex. |
|---|---|
| Source (natural) | Organism:  Azotobacter vinelandii DJ (bacteria) Azotobacter vinelandii DJ (bacteria) |
| Molecular weight | Theoretical: 63 KDa |
-Macromolecule #1: Nitrogenase iron protein gamma chain
| Macromolecule | Name: Nitrogenase iron protein gamma chain / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Azotobacter vinelandii DJ (bacteria) Azotobacter vinelandii DJ (bacteria) |
| Sequence | String: MAMRQCAIYG KGGIGKSTTT QNLVAALAEM GKKVMIVGCD PKADSTRLIL HSKAQNTIME MAAEAGTVE DLELEDVLKA GYGGVKCVES GGPEPGVGCA GRGVITAINF LEEEGAYEDD L DFVFYDVL GDVVCGGFAM PIRENKAQEI YIVCSGEMMA MYAANNISKG ...String: MAMRQCAIYG KGGIGKSTTT QNLVAALAEM GKKVMIVGCD PKADSTRLIL HSKAQNTIME MAAEAGTVE DLELEDVLKA GYGGVKCVES GGPEPGVGCA GRGVITAINF LEEEGAYEDD L DFVFYDVL GDVVCGGFAM PIRENKAQEI YIVCSGEMMA MYAANNISKG IVKYANSGSV RL GGLICNS RNTDREDELI IALANKLGTQ MIHFVPRDNV VQRAEIRRMT VIEYDPKAKQ ADE YRALAR KVVDNKLLVI PNPITMDELE ELLMEFGIME VEDESIVGKT AEEV |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.6 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: Solutions were prepared and filtered immediately prior to the experiment. | |||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING | |||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: HOMEMADE PLUNGER Details: Custom manual plunger. Greater than 95% humidity.. | |||||||||
| Details | 3.6 mg/mL FeP Sample also contained 1.44 mg/mL MoFeP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Temperature | Min: 93.0 K / Max: 123.0 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 14903 / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 135000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: EF / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)