+Search query
-Structure paper
| Title | Structures of the nitrogenase complex prepared under catalytic turnover conditions. |
|---|---|
| Journal, issue, pages | Science, Vol. 377, Issue 6608, Page 865-869, Year 2022 |
| Publish date | Aug 19, 2022 |
 Authors Authors | Hannah L Rutledge / Brian D Cook / Hoang P M Nguyen / Mark A Herzik / F Akif Tezcan /  |
| PubMed Abstract | The enzyme nitrogenase couples adenosine triphosphate (ATP) hydrolysis to the multielectron reduction of atmospheric dinitrogen into ammonia. Despite extensive research, the mechanistic details of ...The enzyme nitrogenase couples adenosine triphosphate (ATP) hydrolysis to the multielectron reduction of atmospheric dinitrogen into ammonia. Despite extensive research, the mechanistic details of ATP-dependent energy transduction and dinitrogen reduction by nitrogenase are not well understood, requiring new strategies to monitor its structural dynamics during catalytic action. Here, we report cryo-electron microscopy structures of the nitrogenase complex prepared under enzymatic turnover conditions. We observe that asymmetry governs all aspects of the nitrogenase mechanism, including ATP hydrolysis, protein-protein interactions, and catalysis. Conformational changes near the catalytic iron-molybdenum cofactor are correlated with the nucleotide-hydrolysis state of the enzyme. |
 External links External links |  Science / Science /  PubMed:35901182 / PubMed:35901182 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 1.91 - 3.01 Å |
| Structure data | EMDB-26756, PDB-7ut6: EMDB-26757, PDB-7ut7:  EMDB-26758: Consensus cryoEM map of Azotobacter vinelandii MoFeP in a 1:1 complex (ATP-bound) with FeP during catalytic N2 reduction  EMDB-26759: Locally refined cryoEM map of Azotobacter vinelandii FeP in a 1:1 complex (ATP-bound) with MoFeP during catalytic N2 reduction EMDB-26760, PDB-7ut8:  EMDB-26761: Consensus cryoEM map of Azotobacter vinelandii MoFeP in a 1:1 complex (ADP/ATP-bound) with FeP during catalytic N2 reduction  EMDB-26762: Locally refined cryoEM map of Azotobacter vinelandii FeP in a 1:1 complex (ADP/ATP-bound) with MoFeP during catalytic N2 reduction EMDB-26763, PDB-7ut9: EMDB-26764, PDB-7uta: EMDB-27639, PDB-8dpn: |
| Chemicals | 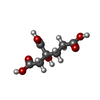 ChemComp-HCA: 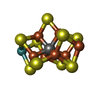 ChemComp-ICS:  ChemComp-FE:  ChemComp-CLF:  ChemComp-HOH:  ChemComp-SF4:  ChemComp-MG:  ChemComp-ATP:  ChemComp-ADP:  ChemComp-0BE: |
| Source |
|
 Keywords Keywords | OXIDOREDUCTASE / nitrogenase / MoFeP / nitrogen fixation / FeP |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers















 azotobacter vinelandii dj (bacteria)
azotobacter vinelandii dj (bacteria)